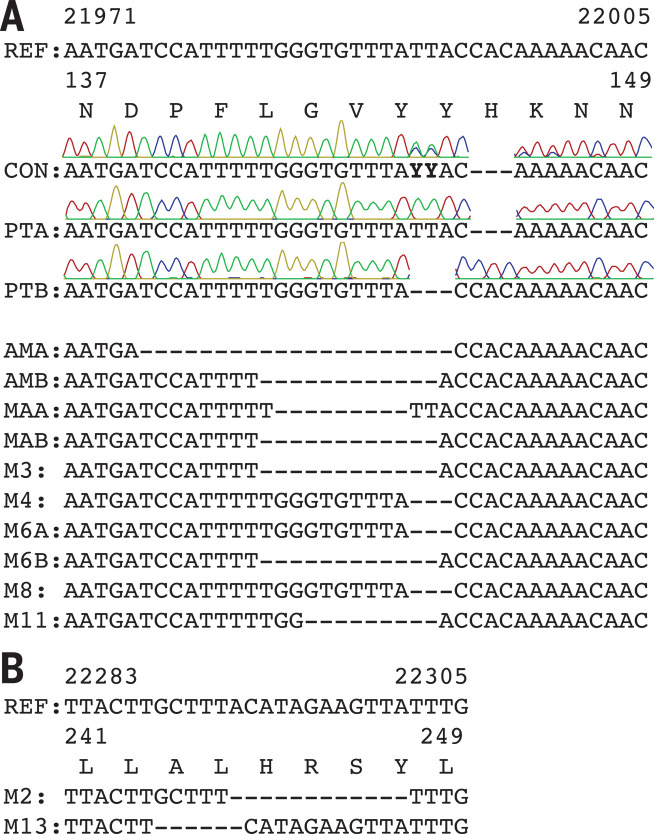
Fig. 1. Deletions in SARS-CoV-2 spike glycoprotein arise during persistent infections of immunosuppressed patients.
(A) Top: Sequences of viruses isolated from PLTI1 (PT) and viruses from patients with deletions in the same NTD region. Chromatograms are shown for sequences from PLTI1, which include sequencing of bulk reverse transcription products (CON) and individual cDNA clones. Bottom: Sequences from other long-term infections from individuals AM (18), MA-JL (MA) (19), and a MSK cohort (M) with individuals 3, 4, 6, 8, and 11 (13). Letters (A and B) designate different variants from the same patient. (B) Sequences of viruses from two patients (M2 and M13) with deletions in a different region of the NTD. All sequences are aligned to reference sequence (REF) MN985325 (WA-1). See fig. S1 for genetic analysis of patient isolates. Amino acid abbreviations: A, Ala; D, Asp; F, Phe; G, Gly; H, His; K, Lys; L, Leu; N, Asn; P, Pro; R, Arg; S, Ser; V, Val; Y, Tyr.