Table 3.
Machine learning and deep learning approaches for plant microbiome-based natural product discovery
|
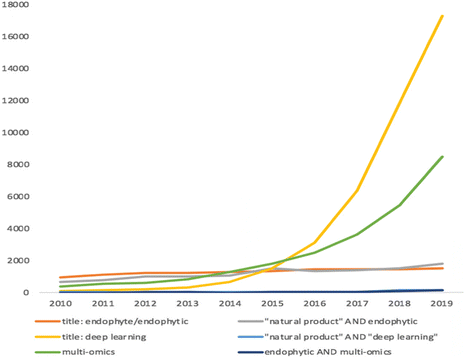
For predicting features of data that are too large to be completely sampled, one of the most promising approaches is computational learning or artificial intelligence, including machine learning and deep learning. These approaches deal with the problem of having an incomplete model to characterize unseen data, by evaluating diverse competing models on a set of training data. In other words, these approaches complete tasks without explicit instructions using patterns (models) learned from the training data. Specific machine learning approaches include Random forests, Hidden Markov Models, hierarchical cluster analyses, and support vector machines. Deep learning is a type of machine learning that handles additional complexity by using layers of data transformations. Specific deep learning approaches use convolutional neural networks where each layer learns from other, previous layers which are called hidden layers. One common framework for building such tools is the well-supported R Interface ‘H2O’ Scalable Machine Learning Platform (GitHub at h2oai/h2o-3) [53]. For global endophyte NP bioprospecting, we can integrate phylogenomic deep learning and genome-wide metabolic model deep learning frameworks. For example, using Pathway Tools v.23.0 [54] integrated with MetaFlux in antiSMASH [55] and DeepBGC [56]. For predicting the chemical structural diversity of endophytes, we can interface the approaches above into chemoinformatic and drug discovery deep learning frameworks. For discovery of in planta unsilencing triggers – waking the sleeping giant, we can integrate experimental system data, OSMAC, and multi-omics data (e.g. from data mining amplicon sequencing, shotgun sequencing, metatranscriptomic sequencing, and metabolomics) Table 1Inset: Recent trends in peer-reviewed studies with keywords/title “endophyte”, “endophyte and natural product”, showing limited increase, whereas studies on “deep learning”, “multi-omics” are steeply increasing |