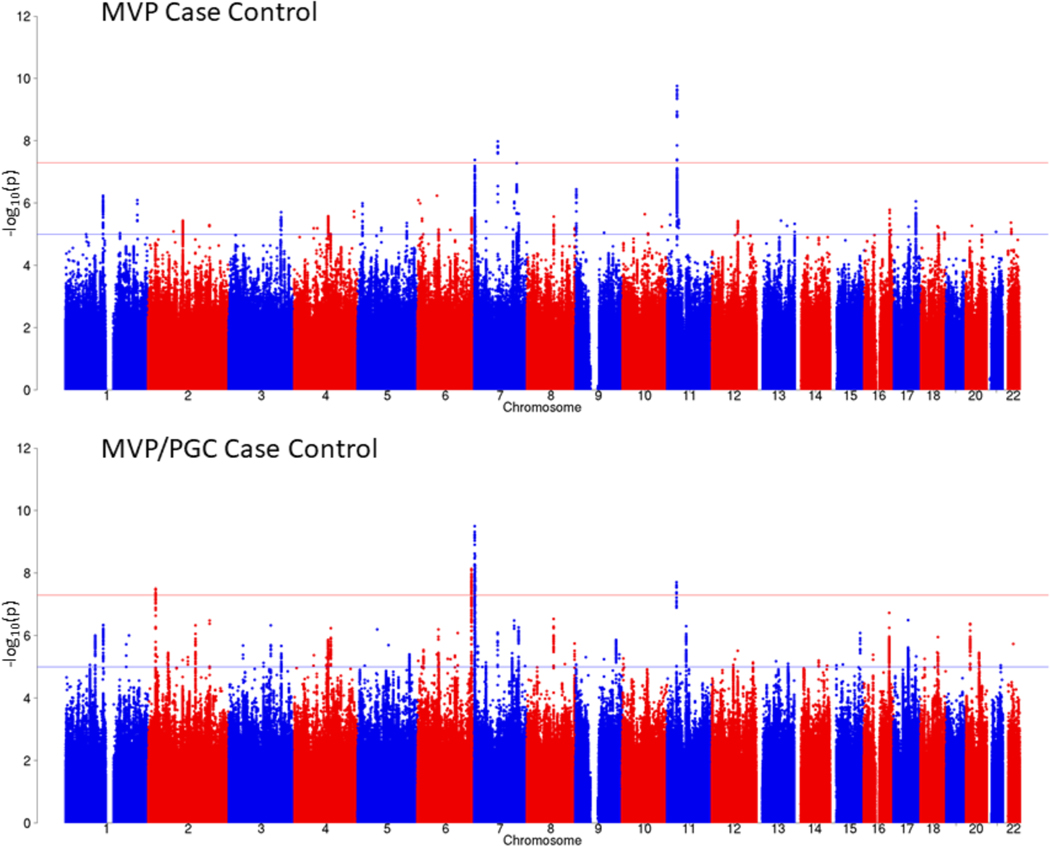
Figure 1 |. Manhattan plot for the MVP case-control GWAS (top) and for the MVP/PGC GWAS meta-analysis in EUR samples (bottom).
GWAS was performed using logistic regression, co-varying for age, sex, and the first 10 principal components of ancestry. Meta-analysis was conducted with METAL25 using the inverse variance weighting method. Bonferroni correction was used to correct for multiple comparisons; associations with P ≤ 5 × 10−8 (indicated by the horizontal red bar) were considered to be genome-wide significant.