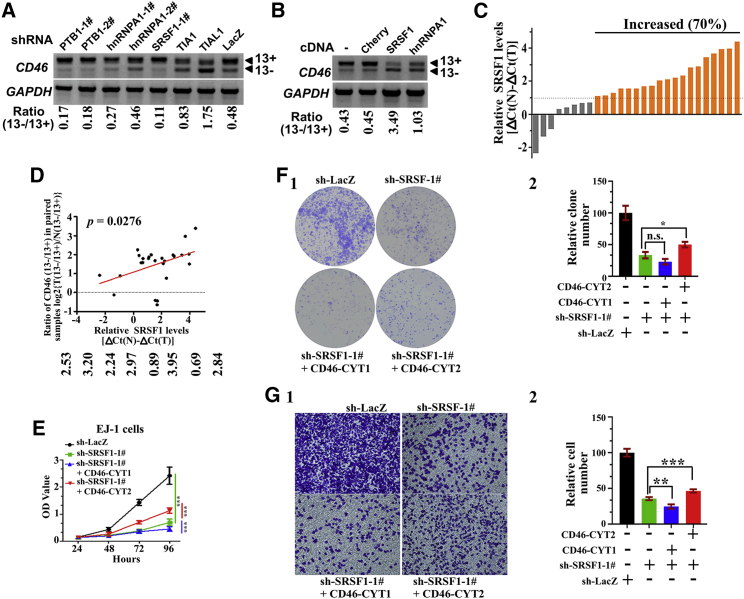
Figure 7.
SRSF1 promotes bladder cancer tumorigenesis in part via regulating CD46-CYT2 levels
(A) EJ-1 cells were infected with lentivirus expressing several indicated shRNAs to establish stably expressing cell lines. Semiquantitative RT-PCR analysis was performed to detect the CD46 exon 13 alternative splicing. The ratio for 13−/13+ is listed below the panel. (B) CD46 exon 13 splicing was measured by RT-PCR in EJ-1 cells stably expressing FLAG-Cherry, FLAG-SRSF1, or FLAG-hnRNPA1. (C) Relative expression of SRSF1 mRNA expression levels were evaluated by real-time PCR in 27 paired case specimens. Expression levels of SRSF1 were normalized to that of GAPDH. A bar value <1 indicates that SRSF1 is decreased in tumors. A bar value >1 indicates that SRSF10 is increased in tumors. (D) The positive correlation between the CD46 13−/13+ ratio and expression levels of SRSF1 was observed in bladder cancer samples. Relationships between these two variables were determined by Pearson’s correlation coefficients. The correlation was analyzed using GraphPad Prism 5 software. (E) A CCK-8 assay was utilized to quantify cell viability at each time point. Data are plotted as the mean ± SD of three independent experiments and were analyzed by two-way ANOVA. ∗∗∗p < 0.001. (F) (1) Representative images of cell culture plates following staining for colony formation of the indicated cell lines. (2) The number of colonies was quantified. The data represent mean ± SD and were analyzed by an unpaired two-tailed Student’s t test (n = 3). ∗p < 0.05. n.s., not significant. (G) (1) Migration assay for the indicated cell lines. (2) Number of migrated cells was quantified in five random images from each treatment group. The data represent mean ± SD and were analyzed by an unpaired two-tailed Student’s t test (n = 5). ∗∗p < 0.01, ∗∗∗p < 0.001.