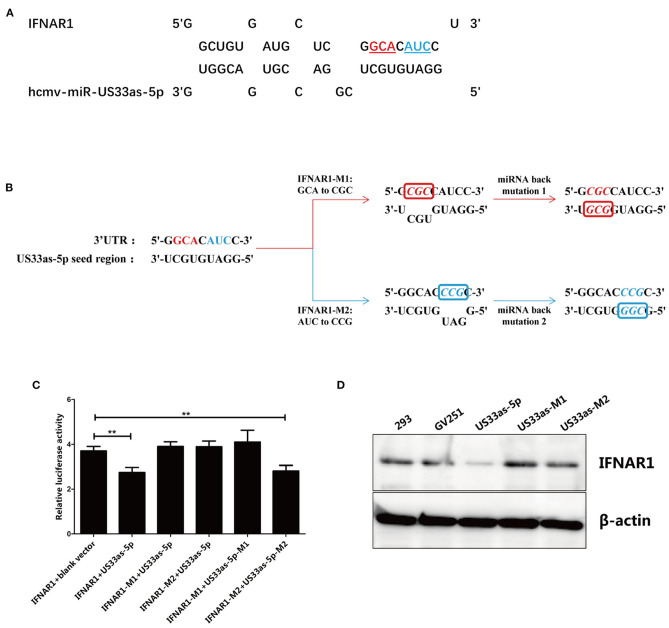
Figure 2.
Main binding site in hcmv-miR-US33as-5p for IFNAR1 mRNA. (A) Schematic of the targeting of IFNAR1 by hcmv-miR-US33as-5p. Mutated nucleotides are shown in color and underlined. (B) Mutation of the IFNAR1-binding site and back mutation of the hcmv-miR-US33as-5p seed region. For IFNAR1 mutagenesis 1, GCA was mutated to CGC, and the miRNA was complementarily mutated to GCG for miR-US33as-5p back mutagenesis 1. For IFNAR1 mutagenesis 2, AUC was mutated to CCG, and the miRNA was complementarily mutated to CGG for miR-US33as-5p back mutagenesis 2. The pmirGLO-IFNAR1-Mutation-UTR vector and GV251-US33as-Mutation-UTR vector were co-transfected into 293 cells cultured in 24-well plates. (C) Dual-luciferase reporter assays to determine relative firefly luciferase activity in 293 cells co-transfected with pmirGLO-IFNAR1-Mutation-UTR vector and GV251-US33as-Mutation-UTR vector were performed, and the calculated data are shown in Figure 1A. (D) Western blot showing the expression of IFNAR1 in 293 cells co-transfected with pmirGLO-IFNAR1(-Mutation)-UTR vector and GV251-US33as(-Mutation)-UTR vector harvested at the same time, with β-actin used as a loading control. The assays were performed in triplicate wells, and data were collected from two different experiments and are represented as the means ± SDs; **p < 0.05.