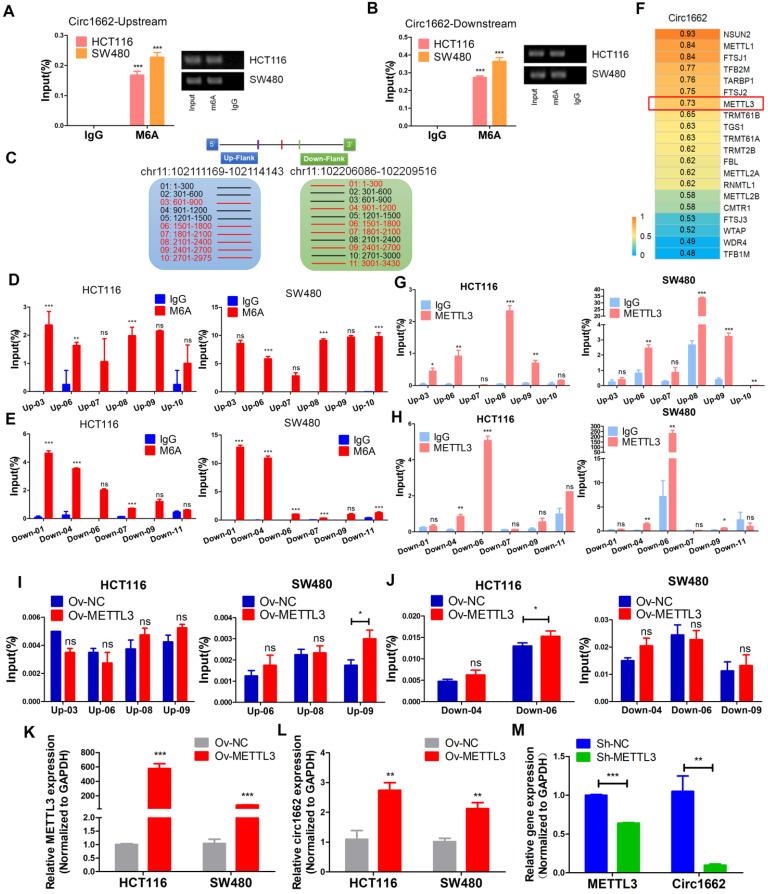
Figure 5.
METTL3 induces circ1662 expression by marking m6A sites in flanking reverse complementary sequences. (A, B) Circ1662 up-and down flank sequence existed m6A modification using MeRIP-qPCR assay. The qPCR production was identified by agarose gel electrophoresis. The result was normalized by input group, IgG group was the negative control. (C) The flank sequences of circ1662 were divided into 300 nt fragments. The circ1662-up-03, -06, -07, -08, -09, and-10 was marked as red fragements which contain reverse complementary sequences. Circ1662-down-01, -04, -06, -07, -09 and -11 was marked as red fragements which contain reverse complementary sequences. (D, E) MeRIP-qPCR assay detecting circ1662-up-03, -06, -07, -08, -09, -10 and circ1662-down-01, -04, -06, -07, -09 in HCT116 and SW480. The result was normalized by input group, IgG group was the negative control. (F) The correlation analysis between RNA methylation-related molecule and circ1662 in 6 pair colorectal cancer tissues. (G, H) METTL3-RIP-qPCR assay pulling circ1662-up-03, -06, -07, -08, -09, -10 and circ1662-down-01, -04, -06, -07, -09, -11 in HCT116 and SW480. The result was normalized by input group, IgG group was the negative control. (I, J) MeRIP-qPCR assay performing to pull down circ1662-up-03, -06, -08, -09 and circ1662-down-04, -06, -09 in overexpressed-METTL3 HCT116 and SW480. The result was normalized by input group, IgG group was the negative control. (K, L) qPCR analysis of METTL3 expression and circ1662 expression in HCT116 and SW480 cells transfected METTL3-overexpressed vector. (M) qPCR analysis of METTL3 expression and circ1662 expression in HCT116 transfected Sh-METTL3 vector. GAPDH is used as an internal reference for performing qPCR. Statistical significance was calculated by Student's t test, *P < 0.05, 0.001 < ** P < 0.01, *** P < 0.001, Mean ± SEM.