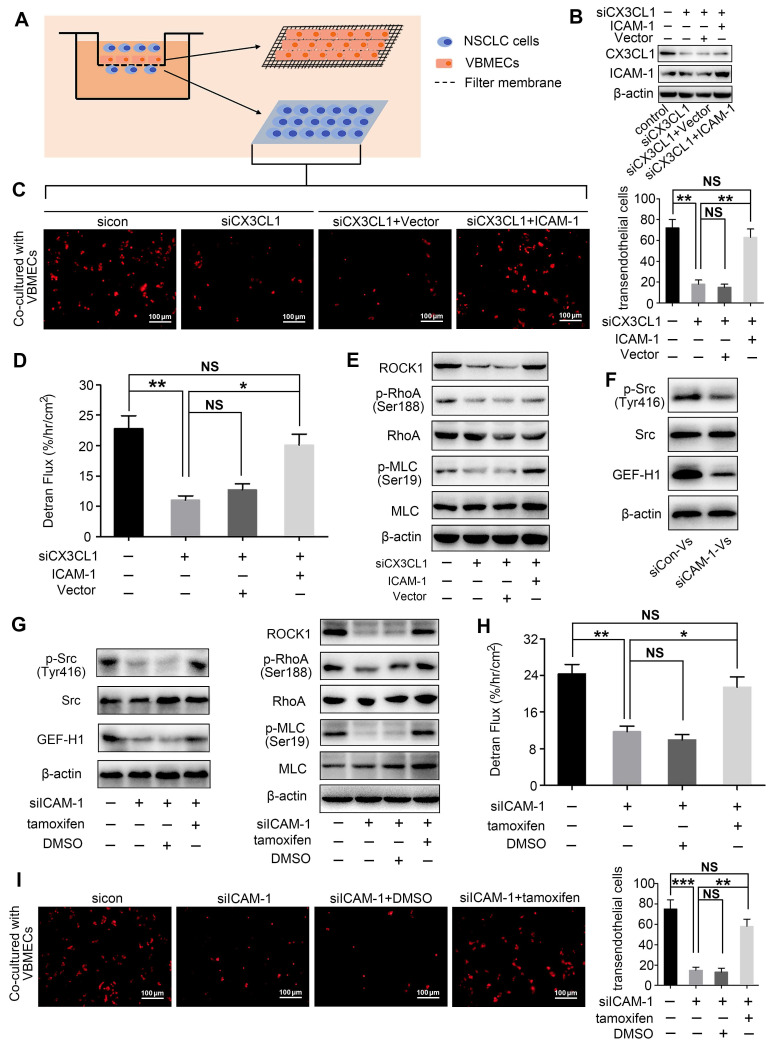
Figure 5.
CX3CL1/ICAM-1 enhanced NSCLC cell transendothelial migration by increasing VBMEC permeability through the Src/GEF-H1 pathway. (A) Schematic illustration of the Transwell analysis of transendothelial migration assays. (B) Western blotting analysis of CX3CL1 and ICAM-1 protein levels in indicated VBMECs. (C) Quantitative analysis of A549 cell transendothelial migration with control (sicon), CX3CL1-KD (siCX3CL1), CX3CL1-KD and Vector (siCX3CL1+Vector) or CX3CL1-KD and ICAM-1-overexpression (siCX3CL1+ICAM-1) VBMECs as an endothelial monolayer barrier. Data represent the mean ± SEM (n = 3). **P < 0.01 and NSP > 0.05. (D) Transport of FITC-labeled dextran through the indicated VBMEC barrier treated with conditioned media from A549 cells. Data represent the mean ± SEM (n = 3). *P < 0.05, **P < 0.01, and NSP > 0.05. (E) Western blotting of RhoA, p-RhoA, MLC, p-MLC, and ROCK1 protein levels in the indicated VBMECs treated with conditioned media from A549 cells. (F) Src, p-Src, and GEF-H1 levels in the indicated VBMECs treated with conditioned media from A549 cells detected by western blotting analysis. (G) Western blotting of RhoA, p-RhoA, MLC, p-MLC, and ROCK1 protein levels in the indicated VBMECs incubated with conditioned media from A549 cells, treated with the Src signaling pathway agonist tamoxifen. (H) Transport of FITC-labeled dextran through the indicated VBMEC barrier induced by conditioned media from A549 cells and treated with tamoxifen. Data represent the mean ± SEM (n = 3). *P < 0.05, **P < 0.01, and NSP > 0.05. (I) Quantitative analysis of NSCLC cell transendothelial migration with administration of tamoxifen in the indicated co-cultures of VBMECs and A549 cells. Data represent the mean ± SEM (n = 3). **P < 0.01, ***P < 0.01, and NSP > 0.05.