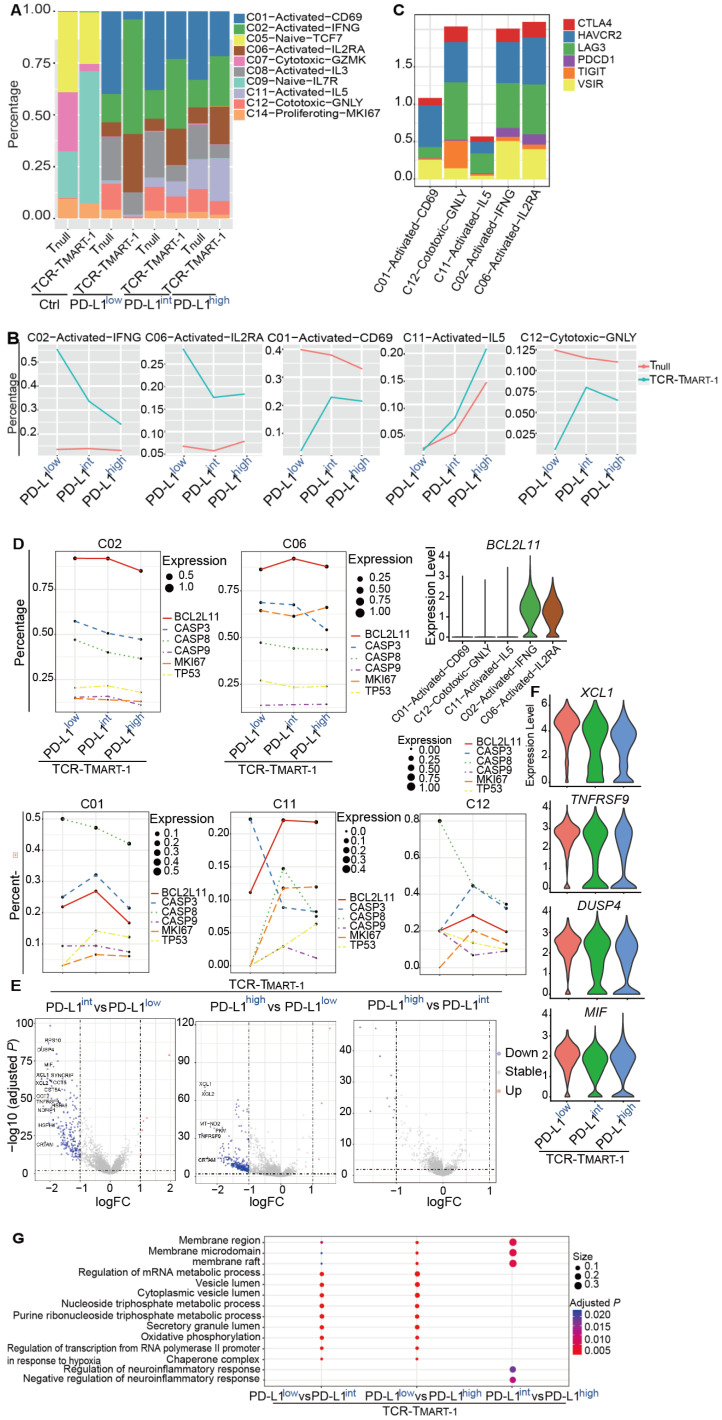
Figure 3.
Various responses between T cell clusters, and Tnull and TCR-TMART-1 to different levels of tumor PD-L1. (A) Cluster composition of Tnull and TCR-TMART-1. (B) The proportion distribution of T cell clusters with the increased tumor PD-L1. (C) The bar plot shows the proportion distribution of cells expressing CTLA4, HAVCR2 (TIM3), LAG3, PDCD1, TIGIT, and VSIR among the five T cell clusters, respectively (cutoff: UMI of the gene > 0). (D) The bubble plot shows the proportion distribution of T cells expressing BCL2L11, CASP3, CASP8, CASP9, MKI67, and TP53 in TCR-TMART-1 responding to differential proportions of PD-L1+ tumor, among the five clusters respectively. The size of the point shows the mean expression of genes in the corresponding T cell population. The violin shows the expression distribution of BCL2L11 among the five clusters. (E) Differentially expressed genes in TCR-TMART-1 responding to differential proportions of PD-L1+ tumor. (F) The expression distribution of XCL1, TNFRSF9, DUSP4, and MIF in TCR-TMART-1 responding to differential proportions of PD-L1+ tumor. (G) Bubble plot showing the top 10 pathways in Tnull (left) and TCR-TMART-1 (right) compared to the control group, respectively. The color represents pvalue and the size represents gene ratio.