Fig. 4. Simulations show that model violations and unequal rates result in artifactual support for monophyletic Deuterostomia.
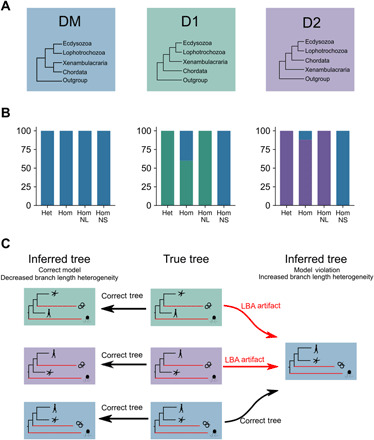
(A) One hundred datasets were simulated using parameters estimated under a site heterogeneous model (CAT + LG + G) for each of the three topologies shown (colored boxes). (B) For each simulated dataset, a maximum likelihood tree was reconstructed under four conditions: i) site heterogeneous C60 + LG + F + G model (Het); ii) model violating site homogeneous LG + F + G model (Hom); iii) homogeneous model with long branches of protostomes and outgroup taxa removed (HomNL); iv) homogeneous model with short-branch protostomes and outgroup taxa removed (HomNS). For each condition, the number of times the three possible topologies were reconstructed is shown in the bar charts; color indicates the topology supported. Data simulated under DM always yield the correct topology. Under the site heterogeneous model, D1 and D2 data always yield a correct topology. Under the model violating site homogeneous model, D1 and D2 data yield an incorrect topology in 40% and 12% of replicates, respectively. The incorrect tree is always DM. Under the site homogeneous model with long-branch protostomes and outgroups removed (reducing LBA), D1 and D2 data always yield a correct topology. Under the site homogeneous model with short-branch protostomes and outgroups removed (enhancing LBA), D1 and D2 data always yield an incorrect topology. The incorrect tree is always DM. (C) Interpretation of simulation results as an LBA artifact. The tree topologies under which data were simulated (true tree) are shown in the middle. The long branches leading to the protostomes and outgroups are indicated in red. Under conditions that minimize systematic error (left, site heterogeneous model or reduction in branch length heterogeneity by removing long branches), the correct tree (DM, D1, or D2) is always reconstructed. Under conditions that enhance systematic error (right, model violating site homogeneous models or increase in branch length heterogeneity by removing short branches), the DM topology is reconstructed using data simulated under all three topologies.
