Fig. 3. Rational design of SARS-CoV-2 spike-presenting NP vaccines.
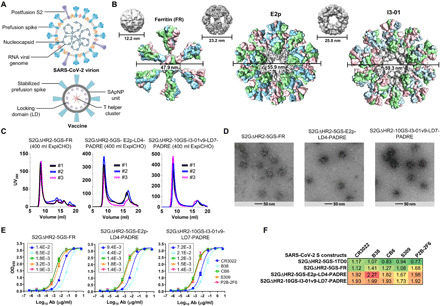
(A) Schematic representation of SARS-CoV-2 virion (top) and spike-presenting SApNP vaccine (bottom). For the SARS-CoV-2 virion, pre-/postfusion S, nucleocapsid, and RNA viral genome are shown, while for the vaccine, stabilized spike and multilayered SApNP composition are depicted. (B) Colored surface models of SApNP carriers (top) and spike-presenting SApNP vaccines (bottom). The three SApNP carriers used here are FR 24-mer and multilayered E2p and I3-01v9 60-mers (the LD layer and PADRE cluster are not shown). Particle size is indicated by diameter (in nanometers). (C) SEC profiles of three SApNPs presenting the SARS-CoV-2 S2GΔHR2 spike, from left to right, S2GΔHR2-5GS-FR, S2GΔHR2-5GS-E2p-LD4-PADRE (or E2p-L4P), and S2GΔHR2-10GS-I3-01v9-LD7-PADRE (or I3-01v9-L7P). SEC profiles were obtained from a Superose 6 10/300 GL column, each showing results from three separate production runs. (D) EM images of three SARS-CoV-2 S2GΔHR2 SApNPs. (E) ELISA binding of three SARS-CoV-2 S2GΔHR2 SApNPs to five mAbs. EC50 (μg/ml) values are labeled. (F) Antigenic profiles of SARS-CoV-2 S2GΔHR2 spike and three SApNPs against five mAbs. Sensorgrams were obtained from an Octet RED96 using six antigen concentrations (150 to 4.6 nM for spike, 9 to 0.27 nM for FR SApNP, and 3.5 to 0.1 nM for E2p and I3-01v9 SApNPs, respectively, all by twofold dilutions) and AHQ biosensors, as shown in fig. S3B. The peak binding signals (in nanometers) at the highest concentration are listed. Color coding indicates the signal strength measured by Octet (green to red: low to high).
