Fig. 1. Tau-targeted ZFP-TFs reduce mouse tau mRNA and protein expression in vitro.
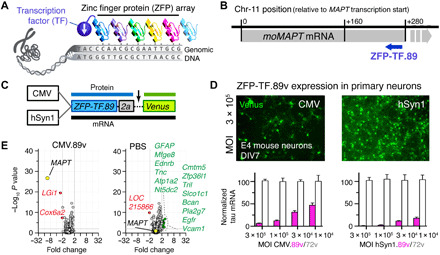
(A) Principle of ZFP-TF target DNA sequence recognition. Engineered arrays of ZFPs recognizing specific DNA triplets bind a specific genomic DNA sequence. Fusion to a TF enables repression of target gene transcription. (B) The binding locations of the lead ZFP-TF.89 downstream of the mouse MAPT gene TSS in the mouse chromosome 11 ZFP-TF.89. Gray bar indicates the location of the MAPT gene. Blue arrow indicates the coding direction of the ZFP-TF.89. (C) Adeno-associated virus serotype 9 (AAV9) constructs with CMV (ubiquitous) and hSyn1 (neuron-specific) promoters for expression of nuclear ZFP-TF.89v (N-terminal SV40 nuclear localization sequence on ZFPs) and cytoplasmic yellow/green fluorescent protein Venus as a transduction marker, separated by a self-cleaving 2a peptide. (D) Expression of AAV ZFP-TF.89v under CMV and hSyn1 promoter in primary cortical mouse neurons (DIV 7; 4 days p.i.). hSyn1-driven 89v expression shows higher tau mRNA repression (pink bars) at the same viral doses (MOI). ZFP-TF.72v, a control ZFP-TF with ZFP array without binding sequence in the mouse genome, shows no tau repression (white bars). Data are presented as means ± SD, n = 3 experiments, and data are normalized to tau mRNA in ZFP-TF.72v–expressing neurons at the highest dose (MOI = 3 × 105). (E) mRNA array data (volcano plots) show small changes in gene expression—other than MAPT—for CMV.89 or PBS compared to CMV.72v-treated primary neurons. Other significantly (>2-fold change, P < 0.01) down-regulated (red) and up-regulated (green) genes are listed. RNA array data are available in table S3. Data are presented as means ± SEM. One-way ANOVA with Sidak’s test, n = 5 to 6 biological replicates.
