Abstract
Advances in regenerative medicine technologies will lead to dramatic changes in how patients in rehabilitation medicine clinics are treated in the upcoming decades. The multidisciplinary field of regenerative medicine is developing new tools for disease modeling and drug discovery based on induced pluripotent stem cells (iPSCs). This approach capitalizes on the idea of personalized medicine by using the patient’s own cells to discover new drugs, increasing the likelihood of a favorable outcome. The search for compounds that can correct disease defects in the culture dish is a conceptual departure from how drug screens were done in the past. This system proposes a closed loop from sample collection from the diseased patient, to in vitro disease model, to drug discovery and FDA approval, to delivering that drug back to the same patient. Here we review recent progress in patient-specific iPSC derivation, directed differentiation toward diseased cell types and how those cells can be used for high throughput drug screens. Given that restoration of normal function is a driving force in rehabilitation medicine, we believe this drug discovery platform focusing on phenotypic rescue will become a key contributor to therapeutic compounds in regenerative rehabilitation.
Harnessing the power of regeneration
In recent years, regenerative medicine technologies have crossed the boundaries of numerous scientific fields, including stem cell, developmental and molecular biology, materials science, engineering, genetics, immunology, physiology and pharmacology. This relatively new field of research has the potential to radically alter the treatment of diseases characterized by the lack of healthy cells or tissues brought on by aging, injury or disease. The goal of regenerative medicine, put simply, is the restoration of function. Restoration is achieved through interventions that either reawaken the body’s endogenous regenerative capacity, or by exogenously supplying normal cells, scaffolds or reconstructed tissues. As one might expect, a combination of these approaches is often the most effective. Regardless, the chosen regenerative intervention must circumvent the default healing pathway, which typically involves inflammation and the deposition of scar tissue, and in its place recapitulate normal tissue architecture that occurs during fetal development. One of the most promising therapeutic approaches proven to achieve or promote regeneration and repair is the transplantation of a wide variety of stem cells. Stem cells are undifferentiated cells that have the capacity for unlimited self-renewal and the ability to differentiate into all of the specialized functional cells of the body. To a large extent, advances in regenerative medicine are contingent on our ability to isolate, propagate and manipulate stem cells. Once transplanted, therapeutic success depends on the cells’ ability to execute their genetic program and interact with their new microenvironment to produce replacement cells de novo, or to elicit repair through paracrine effects. One appealing advantage to this approach is that stem cells offer an unlimited source of material, with the potential to treat diseases that require the replacement of large numbers of cells, like diseases affecting skeletal muscles. Harnessing this powerful regenerative capacity is one of the principle endeavors of regenerative medicine.
Why are stem cells so important?
Different types of stem cells can be classified by their degree of developmental potential. Of the cells that can be cultured in the laboratory, the cells with the most potential are embryonic stem (ES) cells. Derived from the inner cell mass of the blastocyst prior to the germ cell/soma separation [1], they can differentiate into all 200 specialized cells in the body, making them the benchmark of pluripotency. As the embryo develops, almost all of its cells carry out pre-programmed lineage commitment, thereby extinguishing further pluripotentiality. However, a select few stem cells are maintained in the adult in specialized anatomical structures called a niche. These somatic or adult stem cells are normally quiescient and are responsible for day-to-day tissue homeostasis, but can mobilized to repair their organ-of-residence after injury. These somatic stem cells are categorized as being multipotent, in that they can become two or more lineages but are typically restricted to making specialized cells found in their tissue of origin. Hematopoietic stem cells (HSCs) [2][3] were the first example of this category and have an esteemed history in autologous and allogeneic bone marrow transplantation [4, 5]. Mesenchymal stem cells (MSCs), also known as multipotent stromal cells, [6–8] are gaining widespread acceptance as a versatile tool in regenerative medicine. They can be isolated from bone marrow, fat, peripheral blood and Wharton’s jelly in the umbilical cord [9]. Currently, there are more than 125 clinical trials underway worldwide to test the safety and effectiveness of MSC transplantation to treat diseases of bone, cartilage, liver, heart, gastrointestinal tract and the nervous system [10]. MSC’s forte is their ability to home to injured tissue by targeting the inflammatory response, coupled with an ever growing list of reparative functions once they arrive. In various model systems, MSCs have been shown to promote angiogenesis, down-regulate the immune response, recruit and activate surrounding cells via paracrine signaling, produce new extracellular matrix and in rare cases, engraft and differentiate into tissue-specific cells [11]. Skeptics of this work have focused on the observation that MSCs can influence these processes while only transiently present at the site of injury, sometimes only for a matter of hours [12]. Counter arguments typically contend that a transient occupation of the injury site is sufficient to initiate the regenerative cascade. Perhaps the main drawback to the clinical application of adult somatic stem cells is their limited proliferative capacity in culture. Once adapted to growing on plastic, MSCs, for example, will undergo cellular senescence and progressively lose their ability to differentiate [13] after 10–15 passages due to telomere erosion, accumulation of DNA damage and de-repression of the INK4/ARF locus [14]. Despite these shortcomings, MSCs will be a key component in the regenerative arsenal for years to come.
Manufactured stem cells – a revolution in medicine
Here, we discuss the conceptual and practical issues of using induced pluripotent stem (iPS) cells as an in vitro drug screening platform and a tool for delving into disease etiology in ways that could not be done before. As Drs. Ambrosio and Russell advocated in their editorial on the vertical integration of rehabilitation and regenerative medicine, it is imperative that these two fields fuse early during the development of new and innovative therapies for improved speed and efficiency [15]. Therefore, the goal of this review is to justify the marriage of these disciplines by presenting two examples of how the iPS cell-based disease-in-a-dish approach can be used for drug discovery to treat patients commonly seen in rehabilitation clinics. It is strongly purported that therapeutic compounds identified using this personalized medicine approach have a higher likelihood of working in the patient because the patient’s own cells were used to discover the drug.
A Nobel Prize in less than a decade
The discovery that lineage-restricted somatic cells could be dedifferentiated into pluripotent stem cells by the forced expression of a small subset of transcription factors opened a new frontier in the study and treatment of diseases, based on the idea of personalized medicine. In 2006, Takahashi and Yamanaka from Kyoto University, Japan, reprogrammed mouse fibroblasts by introducing retroviruses carrying the Oct4, Sox2, Klf4 and c-Myc genes [16]. What resulted was a population of induced pluripotent stem (iPS) cells that are molecularly and functionally very similar to ES cells, although they are not identical [1, 17]. In 2007, the same techniques were applied to human skin fibroblasts by Yamanaka’s group and the Thomson laboratory at the University of Wisconsin, resulting in the first patient-specific stem cells [18–20]. The derivation of these stem cells was heralded as a way to sidestep ethical and moral concerns involved with the harvesting of ES cells from human embryos, and a means of generating an inexhaustible number of cells for autologous cell replacement therapies. However, several hurdles will need to be overcome before individual patients are offered their own pristine hepatocytes as a therapeutic alternative during terminal liver failure, for example. First, the cost and time of producing personalized iPS cell lines will likely limit their clinical implementation, at least for the foreseeable future. Second, several of the reprogramming factors are oncogenes and the retroviral delivery vehicles can cause deleterious insertional mutagenesis, raising concerns that transplanted cells could cause cancer in the recipients, possibly after a long latency [21]. At least so far, the third challenge is the most daunting—producing functional cell populations containing no undifferentiated cells that would lead to tumors upon transplantation [22]. In the meantime, iPS cells are being used to model diseases while technological advances chip away at these impediments to cellular therapies. This ‘disease-in-a-dish’ approach is currently being adapted to study a wide range of ailments with the goal of uncovering new disease etiology not amenable to study using animal models. The modeling of monogenic disorders has predominated to date, but at least conceptually, nothing should deter investigators from tackling genetically complex diseases in the future.
Heart disease in a dish: using iPS cells to study Duchenne muscular dystrophy (DMD) cardiomyopathy
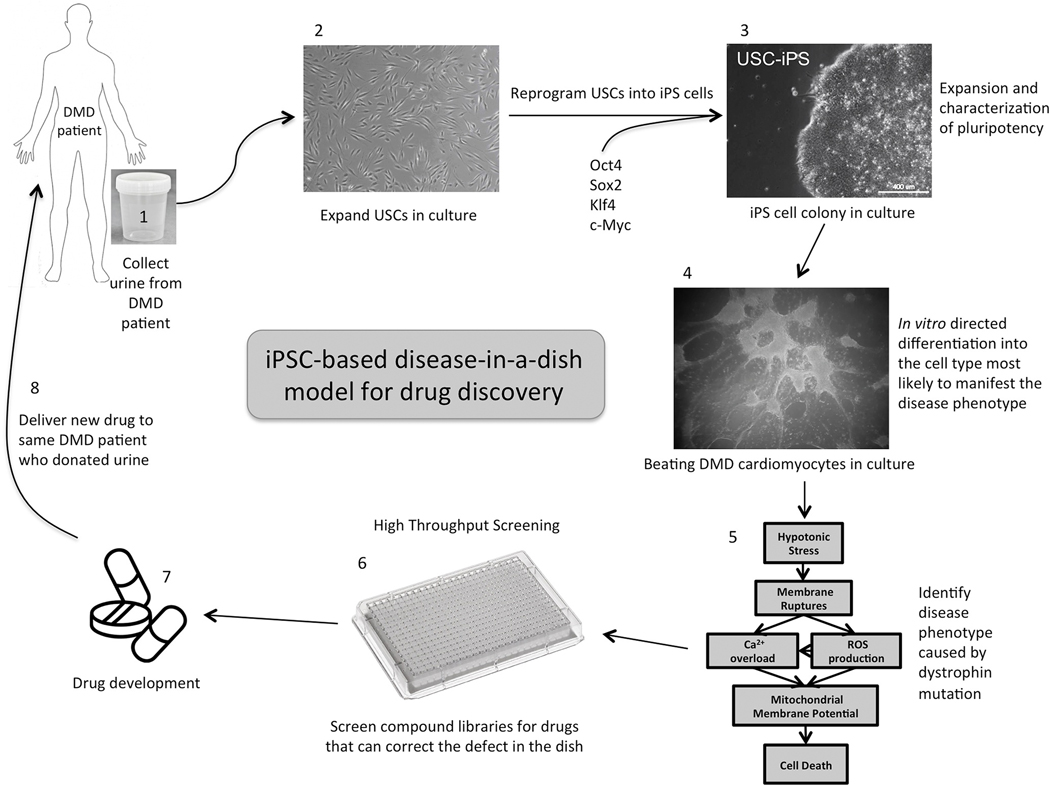
Modeling a disease in culture typically requires differentiating at least one patient-specific iPS cell line into the functional cell type that will mimic as closely as possible the disease phenotype observed in the patient. Our efforts to create an iPS-derived disease model for cardiomyopathy associated with DMD will illustrate the capabilities and shortcomings of the ‘disease in a dish’ approach [23]. To highlight the versatility of this approach and to illustrate specific points, a second iPS model system will be interspersed. That project involves differentiating iPS cells into enteric neurons for the purpose of studying synaptic function in the enteric nervous system in children with Autistic Syndrome Disorder (ASD). A schematic representation of modeling diseases with iPS cells is shown in Figure 1.
Figure 1:
Schematic representation of the sequence of events necessary to model a disease and develop a high throughput drug-screening platform using patient-specific urine-derived iPS cells. The first three steps are the same regardless of the disease being investigated. Steps 4, 5 and 6 will depend on what cell type and defective phenotypic readout make sense to study in your disease of interest.
Issues that need to be considered before embarking on an iPS cell disease-in-a-dish model are summarized in Box 1. DMD-associated cardiomyopathy was chosen as our first foray into this field for several reasons. First, DMD is a monogenic disease, caused by mutations in the dystrophin gene, located on the short arm of the X-chromosome (Xp21.2) and is the largest human gene, covering 2.5 megabases (0.08% of the human genome), spanning 79 exons. Mutations in this gene are typically deletions or duplications, leading to a wide variety of genotype-phenotype relationships, making this disease amendable to the benefits of personalized medicine. The size of the gene makes it a frequent target for mutations, causing DMD to be quite common for a monogenic disease with a frequency of 1:3500 male births. Second, acquiring diseased cardiac tissue to study from young boys with DMD is difficult. Third, the most widely used animal model for DMD, the mdx mouse does not accurately represent the cardiac pathology observed in human patients. Fourth, the molecular mechanism of the cardiomyopathy is not well understood. Early alterations in cellular metabolism and signal transduction associated with a defect in the nitric oxide/cGMP pathway are suspected to be the principle culprits based on animal models. Excessive Ca+2 signaling and the generation of reactive oxygen species with breakdown of the mitochondrial membrane potential have been described in vitro. More recent experiments have shown a link between mitochondrial dysfunction and sarcolemmal injury [24]. Fifth, and most importantly, there is currently no effective treatment available for DMD cardiomyopathy—a condition responsible for early morbidity and mortality in these patients. Onset of the disease typically occurs in early childhood and causes a progressive weakness and wasting of skeletal muscles. DMD cardiomyopathy shows a distinct pattern of myocardial damage, usually detected by echocardiography. Cardiac defects start with diastolic and systolic dysfunction in posterior and lateral wall segments accompanied by arrhythmias, then progress to left ventricular dilation leading eventually to congestive heart failure [25]. Interestingly, there is no correlation between the extent and severity of the skeletal muscle myopathy and the degree and onset of DMD cardiomyopathy, bolstering the need for developing in vitro methods.
Box 1: Issues to consider when modeling diseases with iPS cells for HTS.
Disease Genetics: Monogenic versus polygenic, early versus late onset, degree of penetrance, contribution of environmental factors, single nucleotide polymorphisms versus copy number variants.
Disease Phenotype: Availability of accurate and detailed clinical patient records, severity of phenotype presented, comorbid conditions that may contribute to phenotype.
Choice of control cell line(s): unaffected sibling available or parent, is age- and gender-matching important or will any wild-type iPS cell line suffice?
Selection of somatic cell type to reprogram: fibroblasts, keratinocytes, peripheral mononuclear blood cells, immortalized lymphoblastoid cell lines, urine-derived cells. Cells obtained from repository or first-hand collection from patients? Ease of collection important? Will epigenetic memory of tissue of origin bias ability to differentiate into desired lineage?
Method of reprogramming: retroviral polycistronic vectors containing 4 or 5 transcription factors or non-integrative “footprintless” techniques, stimulus-triggered acquisition of pluripotency (STAP).
Characterization of iPS cell clones: morphology, proliferation, expression of pluripotency markers, teratoma formation in vivo, epigenetic analysis, karyotype. Does the disease impede reprogramming?
Lineage-specific differentiation protocols published? Efficiency, purity and consistency of generating cell type of interest, how well does the protocol recapitulate embryonic development?
Is the disease phenotype cell autonomous? Might other cell types from the same organ work in cooperation to the produce disease phenotype? Is co-culture necessary?
Minimal requirements to confirm that the iPSC-based model retains patient’s genotype and reproduces at least part of the disease phenotype
Can phenotypic assay for the disease be adapted for HTS? Large difference between normal and diseased, must predict clinical outcome, readout should have high signal/noise ratio, wide dynamic range and be automation compatible.
Nervous system disease in a dish: using iPS cells to study autism
Children with ASD often have co-morbid medical conditions [26, 27]. One of the most debilitating of these is chronic gastrointestinal (GI) disturbances, reported to occur in as much as 70% of this population [28–33]. The chronic symptoms are thought to be a result of inflammation, alterations in intestinal permeability, decreased GI motility, abnormal gut microflora, and low-grade endotoxemia [34–39]. A remarkable observation--that successful treatment of GI symptoms is sometimes accompanied by improvements in behavior and cognition in ASD children [40–42]--suggests that the two domains may be linked, however evidence for how or why this occurs is currently lacking. Although there are data showing that defects in the enteric nervous system (ENS) in these children may contribute to their GI symptoms [43–47], the mechanisms have not been adequately described. In contrast, central nervous system (CNS) involvement in ASD is well documented, with multiple studies reporting impaired synaptic development and function that result from mutations in scaffolding proteins, cell adhesion molecules, and genes involved in neuronal signaling pathways [48–56]. For example, mutations and deletions in Shank3, a synaptic scaffolding protein that plays a key role in excitatory/inhibitory balance, are known to be causative in some ASD cases. If patients with ASD have a known mutation/deletion in Shank3 that causes CNS disruption, it is possible that this defect operates at the level of the ENS as well [57–64]. Phelan-McDermid Syndrome (PMS) patients, 85% of whom also have an ASD diagnosis and a approximately 40% who also report chronic GI symptoms, all have a variable size deletion in chromosome 22 that renders them haploinsufficient for Shank3 [65, 66]. Because Shank3 is expressed in both the brain and gut, our goal is to use cells from PMS patients to develop an in vitro model system for the evaluation of synaptic function in the gut.
Will iPS technology work for every disease?
Not all diseases will be equally simple to model and not every aspect of a complex disease will be reproduced in the in vitro model with equal fidelity. This is partly due to the complexity of the genetics, but also dependent on whether the disease phenotype is cell autonomous. If primary cultures can be easily made from the affected tissues by common dissociation techniques, that may be the faster and more cost effective option. In general, the more cell autonomous the phenotype the stronger the argument for developing an iPSC model [67]. Neurological and cardiac syndromes are the best examples and were among the first models exploited for toxicology screening. One of the principle criteria that must be met for the success of an iPSC disease model is having knowledge of, or at least having a strong hunch, what the cellular phenotype is in the patient. That is to say, once the defective cells have been generated in the dish, one must have some idea about what physiological readout to examine. Remember that the overall goal is to produce a clinically relevant readout that recreates the molecular mechanism of pathogenesis. For example, in the iPSC model for spinal muscular atrophy (SMA), neurons produced in the dish showed lower expression of SMN1 and increased neuronal death compared to controls [68] and cardiomyocytes in the long QT syndrome (LQTS) model showed electrical conduction defects nearly identical to those in people with the disease [69]. In our DMD cardiomyopathy model, a wide array of phenotypic characteristics would be predicted from the clinical profile, including defects in Ca2+ handling, mitochondrial function and membrane fragility. Phenotypic differences between dystrophin null cardiomyocytes and normal control cardiomyocytes in each one of these categories was observed [23]. Demonstrating that an iPSC-based model faithfully reproduces the basic phenotypic defect found in the patient would lend credibility to any additional pathological observations made with the model, thus pushing the field forward in a way not previously possible. Although monogenic diseases were attempted first because of the relatively simple relationship between diseased and normal cells, to focus only on Mendelian diseases would sell the iPSC-based models short. Therefore, diseases arising from the complex interplay of multiple mutations and/or secondary modifiers can be modeled effectively, and one could argue that this will be the greatest contribution of this approach because it can be done no other way. In addition, if the disease phenotype in the patient results from the interaction of different cell types, this modeling strategy provides the flexibility to simply differentiate two, or even three different cell types from the same iPS cell clone simultaneously. After each cell type has been characterized individually to the investigators’ satisfaction, they can be co-cultured all together, or in pairwise combinations to investigate the relative role of each cell type in the disease phenotype. Cell type-specific gene knock-down reagents (siRNAs) or small molecule activators or inhibitors could then be employed to provide even more power to address specific predictions. Coupling gene replacement therapy techniques to the iPSC model system would allow “genetic rescue” experiments to be performed. The benefit of doing this is to provide a definitive readout for what a return to normality looks like in the dish so that drug-screening assays have a benchmark for relative effectiveness. In those cases where the disease adversely affects the extent or efficiency of reprogramming, making iPS cells may be impossible without correcting the genetic defect first. Another pitfall arises if the disease is primarily caused by a developmental defect. If one assumes that the most effective and informative differentiation schemes involve recapitulating embryonic development, diseases that halt or obstruct organ development would be particularly problematic to create using iPSC models. Lastly, diseases that have a strong environmental component will probably not be worthwhile to attempt using this technology, unless satisfactory ways can be found to introduce environmental insults.
Where does one start? Choices in cellular reprogramming
What type of somatic cell one chooses to reprogram could vary depending on the disease, age of onset, disease progression, and age of the patient at first contact. iPS cells have successfully been derived from fibroblasts, keratinocytes, mesenchymal stem cells, chord blood cells, peripheral blood mononuclear cells and immortalized lymphoblastoid cell lines—each cell type possessing advantages and disadvantages. Previous studies have shown that cells from young people are more efficiently reprogrammed than those from the elderly. Zhou et al. reported that human urine could be a novel and plentiful source of cells for iPS cell reprogramming [70, 71]. Last year, our laboratory also successfully reprogrammed urine-derived stem cells (USCs) from a DMD patient and a normal volunteer to iPS cells in about two weeks, compared to one month on average for fibroblasts and keratinocytes [23]. The fact that urine can be obtained non-invasively and repeatedly makes it an attractive source of somatic cells for reprogramming. Urine contains a population of somatic stem cells with spindle-shaped morphology and classical mesenchymal stem cell surface markers including CD44, CD73, CD90, CD105 and CD146 [23, 72]. USCs did not express the hematopoietic stem cell (HSC) markers CD25, CD31, CD34 and CD45. In addition, USCs expressed the kidney glomerular podocyte markers podocin and synaptopodin, which is consistent with their mesenchymal origin. Interestingly, USCs isolated for this study showed endogenous expression of two of the reprogramming factors, c-Myc and Klf4 and high telomerase activity, which probably contributed to their faster reprogramming kinetics. USCs are also highly proliferative in culture enabling sufficient quantities of cells for reprogramming to be produced in a short period of time from as little as 50 cc of urine.
Regardless of the somatic cell type chosen for reprogramming, studies have demonstrated that iPS cells are prone to “epigenetic memory” consisting of genome-wide methylation patterns and bivalent histone marks at specific loci [73–75]. Persistence of these genetic adjuncts have been attributed to incomplete removal of somatic cell-specific DNA methylation patterns during reprogramming leading to residual gene expression indicative of their tissue of origin [17, 76]. These observations suggest that certain iPS cell clones will differentiate readily into certain lineages, if that lineage is present in the tissue of origin, but may be difficult or impossible to differentiate into cell types not represented in the tissue of origin. If this “difficult” differentiation is achieved, the resulting cell type could retain a gene expression signature held over from its original tissue, further complicating its characterization. It is widely held that much of the inherent predisposition of any iPS cell clone can be explained by what germ layer (endoderm, mesoderm or ectoderm) gave rise to the tissue of origin. That is to say, it might be difficult to produce pancreatic beta cells (arising from endoderm) from an iPS cell originally derived from keratinocytes (from ectoderm). However, empirical data suggests that these pitfalls may be overcome by “complete” reprogramming and/or the repetitive incubation of iPS cells in inhibitors of DNA methyltransferase activity [77]. It is still not clear whether these observations are a durable characteristic of iPS cells or whether they will eventually be overcome as reprogramming methodologies improve.
Embryogenesis in the laboratory
Reprogramming somatic cells to iPS cells is conceptually simple. The forced expression of a small subset of master transcription factors initiates a cascade of epigenetic erasure leading to chromatin availability for transcription and translation. The details of how this process begins, progresses and eventually stabilizes into an auto-feedback loop of pluripotentiality is currently being elucidated, but is beyond the scope of this review. Most investigators are more concerned with the ultimate success of reprogramming which is highly dependent on the fastidious adherence to accepted practices. The combinatorial nature of events that must happen in sequence for cells to become reprogrammed makes somatic cell de-differentiation a rare event. Consequently, to initiate reprogramming most of the emphasis is placed on understanding the interconnections between cell type, cell number, identity and combination of reprogramming factors, efficiency of transgene delivery, stoichiometry and duration of expression and how putative iPS cells become stably independent of the exogenously-delivered transcription factors [67]. Over time, the field has reached consensus on a universal set of criteria that must be met for an iPS colony to be considered fully reprogrammed. These include an ES cell-like colony morphology, positive staining for alkaline phosphatase (a common ES cell marker), down-regulation (or elimination by excision) of retrovirus- or lentivirus-delivered transcription factors, positive immunohistochemical staining for pluripotency markers (Oct4, Sox2 Nanog, Tra-1–81, SSEA4, Rex1 and telomerase), demethylation of specific promoters as assayed by RT-PCR for those genes, a normal karyotype and differentiation into tissues representing all three germ layers in mouse teratoma formation assays.
Regardless of the type of stem cells, expansion in culture might allow an inherent genomic instability to manifest as deleterious chromosomal rearrangements, leading to spurious results in disease modeling [78–80]. Therefore, it is essential to prove that the iPS cells, and their differentiated counterparts, have not undergone chromosomal aberrations. This is especially important given recent studies showing that iPS cells are prone to higher levels of genetic and epigenetic abnormalities compared to ES cells [74, 81–83]. However, this is not an insurmountable obstacle. Fastidious and frequent karyotyping of each clone to be used has proven effective in keeping this problem under control and would be advisable prior to committing to the time-consuming optimization of a differentiation protocol.
From urine to a beating heart
Differentiating a panel of patient-specific iPS cell clones into the cell type that will manifest the disease phenotype is only possible if a robust differentiation protocol is available. Most protocols depend on the biological activity of growth factors and cytokines to artificially reconstruct the temporal sequence of embryonic regulatory events that push cells down a certain lineage. Even if dependable differentiation protocols exist, extensive optimization for each iPS cell clone will likely be required to maximize efficiency and consistency [84]. One of the main strengths of the iPSC approach is the ability to produce unlimited numbers of diseased differentiated cells, but a significant decrease in efficiency will quickly diminish yield and dramatically increase costs. An alternative way to look at efficiency of differentiation is purity of the final cell population, and assays will need to be developed to distinguish the desired cell type from “everything else.” Heterogeneity can be rendered less important by enriching for the desired population using lineage- and stage-specific iPS reporter cell lines coupled with fluorescence activated cell sorting (FACS) to collect only those cells expressing a fluorescent marker, under the control of a late-lineage promoter. The biggest challenge to enrichment or purification of the desired cell type is eliminating immature cells, often thought to resemble fetal forms of the adult population. In general, the more mature the cells can become the more likely and closely they are to recreate the disease phenotype in the dish. Only the expected combination of expressed genes will suffice as proof of lineage and level of maturity [85, 86].
As an example, our DMD USC-derived iPSCs were exposed to a combination of growth factors, including Activin A, bone morphogenetic protein 4 (BMP4) and dickkopf 1 (DKK1). When supplied in a strict sequence and defined duration of each factor, they work in concert to regulate the Wnt signaling pathway controlling cardiac differentiation [87]. In our hands, this protocol produced sporadic contracting cardiomyocytes in monolayer culture 8–20 days after induction with an efficiency that varied between 40–90%. Extending cardiomyocyte time in culture by several weeks lead to greater maturity as measured by a panel of cardiac lineage markers including sarcomeric α-actinin, cardiac α− and β−myosin heavy chain, as well as membrane localized connexin43. These cultures were also exhibited functional indicators of maturity including spontaneous action potentials characteristic of nodal, ventricular and atrial subtypes [23].
Rehabilitation in a dish
Techniques are being developed to enhance maturity, including differentiating cells in the presence of extracellular matrix in 2D or constructing pseudo-3D organoids, thereby providing both cell-matrix and cell-cell interactions. Matrix nanotopography and substrate stiffness are turning out to be potent regulators of cell shape and gene expression, strongly suggesting that substituting microfabricated growth environments in place of featureless plastic would enhance maturity [88–91]. A natural extension of this idea--subjecting cells at an intermediate stage of differentiation to the mechanical stimuli encountered in their fully differentiated counterparts in an intact tissue—is also effective. That is to say, if one is trying to produce mature skeletal muscle cells, artificially force them to rhythmically and repetitively stretch and contract in a bioreactor for days or weeks to augment maturity. Similarly, recent evidence suggests that several time-honored rehabilitation practices can enhance recovery in certain neuromuscular diseases after stem cell transplantation. For example, neuromuscular stimulation has been shown to improve the therapeutic effects of transplanted muscle stem cells in dystrophic skeletal muscle [92]. This idea of mechanical or electrical stimulation serves as a striking example of how a practice common in clinical rehabilitation has been utilized in cell culture models to great effect, reinforcing the idea of fusing the fields of regenerative and rehabilitation medicine.
Group therapy in a dish
A complementary approach is to co-culture the diseased cells with a cell type with which it cooperates in the intact organism. This approach is being utilized in our attempts to create iPSC-derived enteric neurons from ASD patients. In those experiments iPSCs will be differentiated to the intermediate stage of neural crest cells (NCCs), then co-cultured with rabbit lower esophageal sphincter smooth muscle (LES) cells. This approach has been used successfully to differentiate internal anal sphincter cells [93] and primary enteric progenitor cells into mature enteric neurons [Robert Gilmont, personal communication]. These cross-species experiments have the powerful advantage of distinguishing the relative contribution of the human-derived cells and the rabbit-derived cells to the overall disease phenotype. Species-specific and enteric neuron stage-specific antibodies can then be utilized to determine success of the co-culture in immunocytochemistry experiments.
Physical exam under a microscope
After obtaining the differentiated cell type, the disease phenotype must be characterized. Generalizations are difficult since this will be different for each disease under study and it would be helpful to use the disease’s impact on the patient as the guide to predict in vitro readouts. At this stage, basic questions can also be beneficial, for example, should the readout be at the biochemical level, cellular level or would a functional outcome measure be more informative? Early in the characterization, it is imperative to confirm that the expected perturbation in the patient is indeed present in the cell culture model. For example in our case, the dystrophin mutation leads to an absence of dystophin protein expression in both the young boy donating urine and the cardiomyocytes beating in the dish, thereby satisfying an essential aspect of any disease model. As a general measure of health, do the diseased iPSCs differentiate as efficiently as their normal counterparts, have cell division or movement defects or show signs of increased apoptosis?
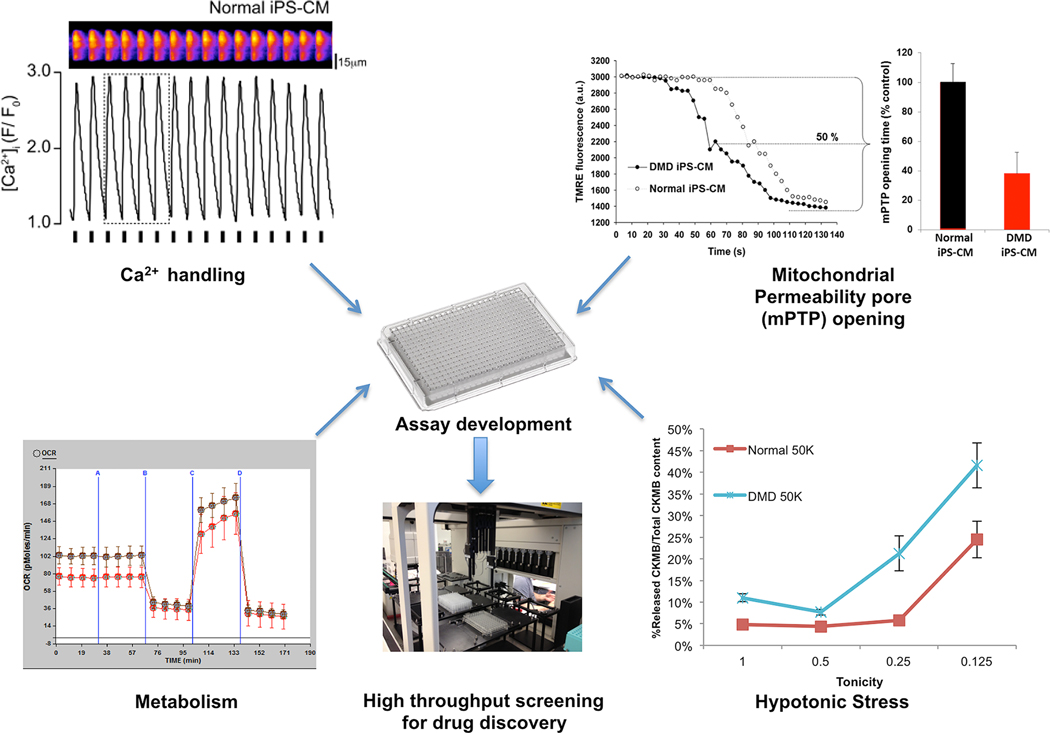
As an illustrative example, our DMD USC-derived beating cardiomyocytes showed defective cardiac function compared to controls in several domains: Calcium handling was impaired with a prolonged recovery time of the calcium transient, mitochondrial permeability pore opening was faster and higher levels of clinically relevant cardiac-specific injury markers (creatine kinase MB and cardiac troponin I [94–97]) were released after hypo-osmotic stress (summarized in Figure 2) [23]. These observations are particularly encouraging since they are consistent with published results on dystrophin’s impact on nitric oxide synthesis and mitochondrial metabolism, as well as its function connecting the cytosolic actin network with the extracellular matrix, thereby protecting the muscle fiber’s sarcolemma from fractures during contractions and ultimately cell death [98–105]. This example also highlights what will be a broadly applicable technique, that is, to challenge the cells in some way that exacerbates the disease phenotype. In our case, a moderate hypo-osmotic stress treatment for 30 minutes accentuated the difference between normal and diseased cells, while serving as a surrogate for years’ worth of contraction-induced injury. It is possible that oxidative stress or catecholamine stimulation could have an indistinguishable affect or bring out a different aspect of this complex phenotype. It remains to be seen whether or not any one of these disease readouts (or a combination of them) can be used as a drug-screening assay looking for the ability of compound X to restore that readout to near normal levels.
Figure 2.
Potential physiological readouts for high-throughput screening using iPS cell-derived dystrophin-null cardiomyocytes. Upper left: Single cell tracing of evoked Ca2+ transients; Upper right: Individual cardiomyocytes derived from normal and DMD were loaded with an inner mitochondria membrane potential dye to measure mitochondria permeability transition pore (mPTP) opening; Lower left: Oxygen consumption rate (OCR) of normal and DMD iPS-CMs measured using the Seahorse™ XF96 Extracellular Flux analyzer; Lower right: Cardiac damage following hypotonic stress. Normal and DMD iPS-CM were incubated in hypotonic solutions and released cardiac troponin I (cTnI) and creatine kinase-MB (CK-MB) were assayed on an enzyme-linked immunosorbent assay (ELISA).
iPSC-based disease models and drug discovery
Historically, animal models, especially mice, were regarded as superior to cell culture-based models for the identification and testing of new drugs. Genetic homogeneity was considered a strong point and the phylogenetic proximity of mice to humans made them the cornerstone of disease modeling for decades. However, mice are not humans. Anatomical, biochemical and physiological divergences between species have derailed thousands of potentially promising drugs, when they work in mice but fail in clinical trials. Differences in drug metabolism, toxicity and penetration of the blood brain barrier are possible explanations for the differential responses in mice and humans. For example, corticosteroids to treat traumatic brain injury showed a clear benefit in mice but failed to show any effect in clinical trials [106]. This unfortunate phenomenon is never more arrestingly evident as when a knock-out mouse has a phenotype closely paralleling the human condition and the compound being tested alleviates the symptoms, only to have the same compound be completely ineffective in patients with that disease. Consequently, there is an enormous unmet need to develop complementary and cost-effective disease models using human cells that reproduce clinically relevant phenotypes with high fidelity, for the purposes of studying disease etiology, toxicology and drug screening.
The list of iPSC-based disease models is steadily growing, with an impressive track record of diseased cells in culture manifesting phenotypes that match symptoms in the patients with those diseases. A partial list includes models for: amyotrophic lateral sclerosis (ALS) [107], spinal muscular atrophy (SMA) [68], familial dysautonomia [108], Rett’s syndrome [109, 110], schizophrenia [111], Parkinson’s disease [112], Timothy syndrome [113, 114] and long QT syndrome (LQTS) [69]. These are listed here to illustrate that many of the mechanistic insights gleaned from these models were facilitated by intricate manipulation of experimental conditions, which is one of the advantages of iPSC-based culture models. Furthermore, because clinically relevant cells are differentiated stepwise from stem cells, in a way that recapitulates embryonic development, it is possible that disease initiation and progression can be observed in the dish. Therefore, it is also possible to exploit these models to uncover early disease markers that appear before overt symptoms in the patient, increasing the opportunity for early intervention, or even prevention. In addition, testing can be done on a single diseased lineage (or a carefully formulated combination of normal and diseased lineages) in isolation, removing it from the secondary effects of residing in a sick animal, which may complicate the analysis. One of the goals of regenerative therapeutics is to target the root cause of the disease and not just alleviate symptoms that might be far removed from the primary molecular defect. Unfortunately, many diseases with the greatest impact on public health might not be amenable to iPSC-based modeling because they are polygenic and/or have a strong environmental component, for example congestive heart failure, diabetes, Alzheimer’s disease and Parkinson’s disease.
Once a dependable phenotypic assay has been identified, adapting the system for high throughput drug screening (HTS) is mostly a matter of scale-up, purity, and reproducibility. Screens of 10,000+ compound library, which is considered a small drug screen, requires an enormous number of highly pure differentiated cells to keep well-to-well variation at an absolute minimum. Developing an HTS-compatible workflow requiring very few manipulations will also go a long way to keeping variation and costs down. Successfully identifying “hits” from large compound libraries may come down whether or not the difference between the normal state and diseased state is large enough to be readily detected in a snap-shot type assay. However, if this is not the case, iPSC-based HTS approaches are also amenable to high-content image-based assays that are able to distinguish and quantify those cells that are positively impacted by a drug candidate and those that are indifferent. Individual wells can be monitored over time and sophisticated microscopy algorithms can be employed to pick up more subtle differences. Another important consideration is the determination of what phenotypic rescue would look like, so the HTS assay can recognize it when it happens. This is where coupling iPSC models with gene therapy moves to the forefront. For monogenic diseases, introducing the wild-type copy of the gene will produce phenotypic rescue in a genetically identical cell type that is at the same stage developmentally because both were differentiated using the same protocol, thereby minimizing variation from differences in genetic background between patients. For example, experiments currently underway in our laboratory are designed to determine whether restored dystrophin expression driven by a hybrid cardiac-specific promoter [115] is sufficient to rescue function in any or all of the domains described above.
Once a candidate small molecule passes muster in the iPSC-based HTS, an appropriate animal model will be needed to validate these “hits”. However, if the compound identified is already an Food and Drug Administration (FDA)-approved drug, with known pharmacokinetic and toxicity profiles, it may be the case that no additional animal studies are needed. For molecules not yet approved, it is our contention that iPSC-based in vitro models and pre-existing mouse models should work in concert to pick better candidate drugs before venturing into the incredibly expensive realm of a clinical trial. Furthermore, during the FDA approval process, that can take many years, iPSC-based models are inherently more nimble and can be modified quickly with great precision to address issues that might arise.
An alternative to high cost drug discovery
It costs about $1.2 billion to bring a single new drug to market in the US today [116]. This exorbitant cost can be attributed partly to the ever-rising price of basic research and development and pre-clinical animal studies, but mostly to the protracted and antiquated path through clinical trials. This crippling combination, coupled with an ever-growing number of late stage failures has caused the pipeline to drop to an all time low [117] and made the drug development process in our country unsustainable. This unfortunate situation has forced drug companies to focus almost exclusively on potential blockbuster drugs and leaving thousands of promising compounds untested [118]. We believe that iPSC-based in vitro models could counter this disturbing trend by decreasing late-stage failure rates. By synergizing these two complementary approaches more insightful testing could be performed as drugs transition from well-accepted animal models into testing on human cells exhibiting the disease under study.
The greatest unrealized promise of any patient-specific iPSC-based drug discovery approach is that it will be better at finding a drug that works for you. The idea of personalized medicine is moving full steam ahead in the area of comprehensive genomic information but we are still doing population based pharmacology [119]. Everyday in this country, thousands of prescriptions are administered to, or taken by patients in whom that drug has no real chance of working. For decades, pharmaceutical companies have developed drugs based on their ability to interact with a certain signal transduction pathway target, proven to be pathological in most people with a particular disease or ailment. One of the hallmarks of a patient-specific iPSC-based drug discovery approach is that a compound is tested for its ability to correct, or partially alleviate the defect at the phenotypic level, without bias toward a predetermined mechanism that may or may not be relevant in a particular individual.
Rehabilitation medicine and disease in a dish: a new paradigm of regenerative rehabilitation
iPS cell reprogramming and in vitro directed differentiation to physiologically relevant cell types are in their infancy. Given the enormous potential and vast resources now being dedicated to all aspects of iPS cell technology, confidence is high that the majority of the hurdles discussed in this review will be overcome in a relatively short period of time. For example, reprogramming methods are being developed that avoid oncogenes and that employ either excisable viral vectors, or episomal vectors that eliminate the possibility of genomic integration [120–122]. In fact, a recently published paper by Obokata et al. reported that splenic CD45+ lymphocytes can be reprogrammed to iPS cells by simply exposing them to pH=5.7 for 25 minutes [123]. This observation alone stands to revolutionize yet again how patient-specific stem cells can be generated. At the very least, ongoing research into the mechanisms of stem cell pluripotency and their differentiation into functional cell types will improve our understanding of regenerative processes and lead to safer and more effective treatments. It has also been suggested that complete reprogramming, i.e., restoring ES-equivalent pluripotentiality, may not be the most pragmatically useful approach. Partial reprogramming to a stage that will readily differentiate into the cell type of interest, while not being teratoma-forming, might be a faster, safer and more economically viable alternative. Whether or not iPS cells will completely replace ES cells remains to be seen, but it is becoming clear that no single type of stem cell will fit the wide range of applications under development. It is too early to come to a definitive conclusion about whether iPS cell technology will make clinically important contributions to disease modeling and drug discovery, but as advances are made, the hope is that innovative therapies arising from this technology will quickly find its way into clinical rehabilitation treatment paradigms.
References:
- 1.Puri MC and Nagy A, Concise review: Embryonic stem cells versus induced pluripotent stem cells: the game is on. Stem Cells, 2012. 30(1): p. 10–4. [DOI] [PubMed] [Google Scholar]
- 2.Ogawa M, Differentiation and proliferation of hematopoietic stem cells. Blood, 1993. 81(11): p. 2844–53. [PubMed] [Google Scholar]
- 3.Seita J. and Weissman IL, Hematopoietic stem cell: self-renewal versus differentiation. Wiley Interdiscip Rev Syst Biol Med, 2010. 2(6): p. 640–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas ED, Bone marrow transplantation from the personal viewpoint. Int J Hematol, 2005. 81(2): p. 89–93. [DOI] [PubMed] [Google Scholar]
- 5.Vaughan W, et al. , The principles and overview of autologous hematopoietic stem cell transplantation. Cancer Treat Res, 2009. 144: p. 23–45. [DOI] [PubMed] [Google Scholar]
- 6.Prockop DJ, Repair of tissues by adult stem/progenitor cells (MSCs): controversies, myths, and changing paradigms. Mol Ther, 2009. 17(6): p. 939–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caplan AI, Why are MSCs therapeutic? New data: new insight. J Pathol, 2009. 217(2): p. 318–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner J, et al. , Optimizing mesenchymal stem cell-based therapeutics. Curr Opin Biotechnol, 2009. 20(5): p. 531–6. [DOI] [PubMed] [Google Scholar]
- 9.Hass R, et al. , Different populations and sources of human mesenchymal stem cells (MSC): A comparison of adult and neonatal tissue-derived MSC. Cell Commun Signal, 2011. 9: p. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trounson A, et al. , Clinical trials for stem cell therapies. BMC Med, 2011. 9: p. 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karp JM and Leng Teo GS, Mesenchymal stem cell homing: the devil is in the details. Cell Stem Cell, 2009. 4(3): p. 206–16. [DOI] [PubMed] [Google Scholar]
- 12.Kean TJ, et al. , MSCs: Delivery Routes and Engraftment, Cell-Targeting Strategies, and Immune Modulation. Stem Cells Int, 2013. 2013: p. 732742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Digirolamo CM, et al. , Propagation and senescence of human marrow stromal cells in culture: a simple colony-forming assay identifies samples with the greatest potential to propagate and differentiate. Br J Haematol, 1999. 107(2): p. 275–81. [DOI] [PubMed] [Google Scholar]
- 14.Estrada JC, et al. , Human mesenchymal stem cell-replicative senescence and oxidative stress are closely linked to aneuploidy. Cell Death Dis, 2013. 4: p. e691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ambrosio F. and Russell A, Regenerative rehabilitation: a call to action. J Rehabil Res Dev, 2010. 47(3): p. xi–xv. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi K. and Yamanaka S, Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell, 2006. 126(4): p. 663–76. [DOI] [PubMed] [Google Scholar]
- 17.Doi A, et al. , Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet, 2009. 41(12): p. 1350–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi K, et al. , Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell, 2007. 131(5): p. 861–72. [DOI] [PubMed] [Google Scholar]
- 19.Yu J, et al. , Induced pluripotent stem cell lines derived from human somatic cells. Science, 2007. 318(5858): p. 1917–20. [DOI] [PubMed] [Google Scholar]
- 20.Park IH, et al. , Reprogramming of human somatic cells to pluripotency with defined factors. Nature, 2008. 451(7175): p. 141–6. [DOI] [PubMed] [Google Scholar]
- 21.Hacein-Bey-Abina S, et al. , LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science, 2003. 302(5644): p. 415–9. [DOI] [PubMed] [Google Scholar]
- 22.Cyranoski D, Stem cells: 5 things to know before jumping on the iPS bandwagon. Nature, 2008. 452(7186): p. 406–8. [DOI] [PubMed] [Google Scholar]
- 23.Guan X, et al. , Dystrophin-deficient cardiomyocytes derived from human urine: New biologic reagents for drug discovery. Stem Cell Res, 2013. 12(2): p. 467–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burelle Y, et al. , Alterations in mitochondrial function as a harbinger of cardiomyopathy: lessons from the dystrophic heart. J Mol Cell Cardiol, 2010. 48(2): p. 310–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yilmaz A. and Sechtem U, Cardiac involvement in muscular dystrophy: advances in diagnosis and therapy. Heart, 2012. 98(5): p. 420–9. [DOI] [PubMed] [Google Scholar]
- 26.Horvath K, et al. , Gastrointestinal abnormalities in children with autistic disorder. J Pediatr, 1999. 135(5): p. 559–63. [DOI] [PubMed] [Google Scholar]
- 27.Dawson G, et al. , Randomized, controlled trial of an intervention for toddlers with autism: the Early Start Denver Model. Pediatrics, 2010. 125(1): p. e17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohane IS, et al. , The co-morbidity burden of children and young adults with autism spectrum disorders. PLoS One, 2012. 7(4): p. e33224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ming X, et al. , Autism spectrum disorders and identified toxic land fills: co-occurrence across States. Environ Health Insights, 2008. 2: p. 55–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valicenti-McDermott M, et al. , Frequency of gastrointestinal symptoms in children with autistic spectrum disorders and association with family history of autoimmune disease. J Dev Behav Pediatr, 2006. 27(2 Suppl): p. S128–36. [DOI] [PubMed] [Google Scholar]
- 31.Buie T, et al. , Evaluation, diagnosis, and treatment of gastrointestinal disorders in individuals with ASDs: a consensus report. Pediatrics, 2010. 125 Suppl 1: p. S1–18. [DOI] [PubMed] [Google Scholar]
- 32.Wang LW, Tancredi DJ, and Thomas DW, The prevalence of gastrointestinal problems in children across the United States with autism spectrum disorders from families with multiple affected members. J Dev Behav Pediatr, 2011. 32(5): p. 351–60. [DOI] [PubMed] [Google Scholar]
- 33.Chaidez V, Hansen RL, and Hertz-Picciotto I, Gastrointestinal Problems in Children with Autism, Developmental Delays or Typical Development. J Autism Dev Disord, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ibrahim SH, et al. , Incidence of gastrointestinal symptoms in children with autism: a population-based study. Pediatrics, 2009. 124(2): p. 680–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Magistris L, et al. , Alterations of the intestinal barrier in patients with autism spectrum disorders and in their first-degree relatives. J Pediatr Gastroenterol Nutr, 2010. 51(4): p. 418–24. [DOI] [PubMed] [Google Scholar]
- 36.Finegold SM, Desulfovibrio species are potentially important in regressive autism. Med Hypotheses, 2011. 77(2): p. 270–4. [DOI] [PubMed] [Google Scholar]
- 37.Finegold SM, et al. , Gastrointestinal microflora studies in late-onset autism. Clin Infect Dis, 2002. 35(Suppl 1): p. S6–S16. [DOI] [PubMed] [Google Scholar]
- 38.Buie T, et al. , Recommendations for evaluation and treatment of common gastrointestinal problems in children with ASDs. Pediatrics, 2010. 125 Suppl 1: p. S19–29. [DOI] [PubMed] [Google Scholar]
- 39.Emanuele E, et al. , Low-grade endotoxemia in patients with severe autism. Neurosci Lett, 2010. 471(3): p. 162–5. [DOI] [PubMed] [Google Scholar]
- 40.Adams JB, et al. , Gastrointestinal flora and gastrointestinal status in children with autism--comparisons to typical children and correlation with autism severity. BMC Gastroenterol, 2011. 11: p. 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pennesi CM and Klein LC, Effectiveness of the gluten-free, casein-free diet for children diagnosed with autism spectrum disorder: based on parental report. Nutr Neurosci, 2012. 15(2): p. 85–91. [DOI] [PubMed] [Google Scholar]
- 42.Sandler RH, et al. , Short-term benefit from oral vancomycin treatment of regressive-onset autism. J Child Neurol, 2000. 15(7): p. 429–35. [DOI] [PubMed] [Google Scholar]
- 43.Anderson RB, Stewart AL, and Young HM, Phenotypes of neural-crest-derived cells in vagal and sacral pathways. Cell Tissue Res, 2006. 323(1): p. 11–25. [DOI] [PubMed] [Google Scholar]
- 44.Berthoud HR, et al. , Neuroanatomy of extrinsic afferents supplying the gastrointestinal tract. Neurogastroenterol Motil, 2004. 16 Suppl 1: p. 28–33. [DOI] [PubMed] [Google Scholar]
- 45.Sanders KM, Koh SD, and Ward SM, Interstitial cells of cajal as pacemakers in the gastrointestinal tract. Annu Rev Physiol, 2006. 68: p. 307–43. [DOI] [PubMed] [Google Scholar]
- 46.Taylor CT and Keely SJ, The autonomic nervous system and inflammatory bowel disease. Auton Neurosci, 2007. 133(1): p. 104–14. [DOI] [PubMed] [Google Scholar]
- 47.Torrente F, et al. , Small intestinal enteropathy with epithelial IgG and complement deposition in children with regressive autism. Mol Psychiatry, 2002. 7(4): p. 375–82, 334. [DOI] [PubMed] [Google Scholar]
- 48.Arons MH, et al. , Autism-associated mutations in ProSAP2/Shank3 impair synaptic transmission and neurexin-neuroligin-mediated transsynaptic signaling. J Neurosci, 2012. 32(43): p. 14966–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baudouin SJ, et al. , Shared synaptic pathophysiology in syndromic and nonsyndromic rodent models of autism. Science, 2012. 338(6103): p. 128–32. [DOI] [PubMed] [Google Scholar]
- 50.Chubykin AA, et al. , Dissection of synapse induction by neuroligins: effect of a neuroligin mutation associated with autism. J Biol Chem, 2005. 280(23): p. 22365–74. [DOI] [PubMed] [Google Scholar]
- 51.Durand CM, et al. , SHANK3 mutations identified in autism lead to modification of dendritic spine morphology via an actin-dependent mechanism. Mol Psychiatry, 2012. 17(1): p. 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gilman SR, et al. , Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron, 2011. 70(5): p. 898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hussman JP, et al. , A noise-reduction GWAS analysis implicates altered regulation of neurite outgrowth and guidance in autism. Mol Autism, 2011. 2(1): p. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moessner R, et al. , Contribution of SHANK3 mutations to autism spectrum disorder. Am J Hum Genet, 2007. 81(6): p. 1289–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.State MW and Levitt P, The conundrums of understanding genetic risks for autism spectrum disorders. Nat Neurosci, 2011. 14(12): p. 1499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sudhof TC, Neuroligins and neurexins link synaptic function to cognitive disease. Nature, 2008. 455(7215): p. 903–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anderlid BM, et al. , FISH-mapping of a 100-kb terminal 22q13 deletion. Hum Genet, 2002. 110(5): p. 439–43. [DOI] [PubMed] [Google Scholar]
- 58.Betancur C. and Buxbaum JD, SHANK3 haploinsufficiency: a “common” but underdiagnosed highly penetrant monogenic cause of autism spectrum disorders. Mol Autism, 2013. 4(1): p. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boccuto L, et al. , Prevalence of SHANK3 variants in patients with different subtypes of autism spectrum disorders. Eur J Hum Genet, 2013. 21(3): p. 310–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bonaglia MC, et al. , Identification of a recurrent breakpoint within the SHANK3 gene in the 22q13.3 deletion syndrome. J Med Genet, 2006. 43(10): p. 822–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Durand CM, et al. , Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet, 2007. 39(1): p. 25–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gauthier J, et al. , Novel de novo SHANK3 mutation in autistic patients. Am J Med Genet B Neuropsychiatr Genet, 2009. 150B(3): p. 421–4. [DOI] [PubMed] [Google Scholar]
- 63.Schaaf CP, et al. , Oligogenic heterozygosity in individuals with high-functioning autism spectrum disorders. Hum Mol Genet, 2011. 20(17): p. 3366–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilson HL, et al. , Molecular characterisation of the 22q13 deletion syndrome supports the role of haploinsufficiency of SHANK3/PROSAP2 in the major neurological symptoms. J Med Genet, 2003. 40(8): p. 575–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Phelan K. and McDermid HE, The 22q13.3 Deletion Syndrome (Phelan-McDermid Syndrome). Mol Syndromol, 2012. 2(3–5): p. 186–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Soorya L, et al. , Prospective investigation of autism and genotype-phenotype correlations in 22q13 deletion syndrome and SHANK3 deficiency. Mol Autism, 2013. 4(1): p. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tiscornia G, Vivas EL, and Izpisua Belmonte JC, Diseases in a dish: modeling human genetic disorders using induced pluripotent cells. Nat Med, 2011. 17(12): p. 1570–6. [DOI] [PubMed] [Google Scholar]
- 68.Ebert AD, et al. , Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature, 2009. 457(7227): p. 277–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Itzhaki I, et al. , Modelling the long QT syndrome with induced pluripotent stem cells. Nature, 2011. 471(7337): p. 225–9. [DOI] [PubMed] [Google Scholar]
- 70.Zhou T, et al. , Generation of human induced pluripotent stem cells from urine samples. Nat Protoc, 2012. 7(12): p. 2080–9. [DOI] [PubMed] [Google Scholar]
- 71.Zhou T, et al. , Generation of induced pluripotent stem cells from urine. J Am Soc Nephrol, 2011. 22(7): p. 1221–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Y, et al. , Urine derived cells are a potential source for urological tissue reconstruction. J Urol, 2008. 180(5): p. 2226–33. [DOI] [PubMed] [Google Scholar]
- 73.Guenther MG, et al. , Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell, 2010. 7(2): p. 249–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lister R, et al. , Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature, 2011. 471(7336): p. 68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meissner A, Epigenetic modifications in pluripotent and differentiated cells. Nat Biotechnol, 2010. 28(10): p. 1079–88. [DOI] [PubMed] [Google Scholar]
- 76.Kim K, et al. , Epigenetic memory in induced pluripotent stem cells. Nature, 2010. 467(7313): p. 285–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Polo JM, et al. , Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol, 2010. 28(8): p. 848–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li H, et al. , Spontaneous expression of embryonic factors and p53 point mutations in aged mesenchymal stem cells: a model of age-related tumorigenesis in mice. Cancer Res, 2007. 67(22): p. 10889–98. [DOI] [PubMed] [Google Scholar]
- 79.Bork S, et al. , DNA methylation pattern changes upon long-term culture and aging of human mesenchymal stromal cells. Aging Cell, 2010. 9(1): p. 54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mayshar Y, et al. , Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell, 2010. 7(4): p. 521–31. [DOI] [PubMed] [Google Scholar]
- 81.Gore A, et al. , Somatic coding mutations in human induced pluripotent stem cells. Nature, 2011. 471(7336): p. 63–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hussein SM, et al. , Copy number variation and selection during reprogramming to pluripotency. Nature, 2011. 471(7336): p. 58–62. [DOI] [PubMed] [Google Scholar]
- 83.Laurent LC, et al. , Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell, 2011. 8(1): p. 106–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Osafune K, et al. , Marked differences in differentiation propensity among human embryonic stem cell lines. Nat Biotechnol, 2008. 26(3): p. 313–5. [DOI] [PubMed] [Google Scholar]
- 85.Irion S, et al. , Directed differentiation of pluripotent stem cells: from developmental biology to therapeutic applications. Cold Spring Harb Symp Quant Biol, 2008. 73: p. 101–10. [DOI] [PubMed] [Google Scholar]
- 86.Murry CE and Keller G, Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell, 2008. 132(4): p. 661–80. [DOI] [PubMed] [Google Scholar]
- 87.Laflamme MA, et al. , Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol, 2007. 25(9): p. 1015–24. [DOI] [PubMed] [Google Scholar]
- 88.Kim DH, et al. , Matrix nanotopography as a regulator of cell function. J Cell Biol, 2012. 197(3): p. 351–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim HN, et al. , Nanotopography-guided tissue engineering and regenerative medicine. Adv Drug Deliv Rev, 2013. 65(4): p. 536–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kshitiz, et al. , Matrix rigidity controls endothelial differentiation and morphogenesis of cardiac precursors. Sci Signal, 2012. 5(227): p. ra41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kshitiz, et al. , Control of stem cell fate and function by engineering physical microenvironments. Integr Biol (Camb), 2012. 4(9): p. 1008–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Distefano G, et al. , Neuromuscular electrical stimulation as a method to maximize the beneficial effects of muscle stem cells transplanted into dystrophic skeletal muscle. PLoS One, 2013. 8(3): p. e54922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Raghavan S, et al. , Successful implantation of bioengineered, intrinsically innervated, human internal anal sphincter. Gastroenterology, 2011. 141(1): p. 310–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Adams JE 3rd, et al. , Cardiac troponin I. A marker with high specificity for cardiac injury. Circulation, 1993. 88(1): p. 101–6. [DOI] [PubMed] [Google Scholar]
- 95.Hoogerwaard EM, et al. , Troponin T and troponin I in carriers of Duchenne and Becker muscular dystrophy with cardiac involvement. Clin Chem, 2001. 47(5): p. 962–3. [PubMed] [Google Scholar]
- 96.Ramaciotti C, Iannaccone ST, and Scott WA, Myocardial cell damage in Duchenne muscular dystrophy. Pediatr Cardiol, 2003. 24(5): p. 503–6. [DOI] [PubMed] [Google Scholar]
- 97.Townsend D, et al. , Chronic administration of membrane sealant prevents severe cardiac injury and ventricular dilatation in dystrophic dogs. J Clin Invest, 2010. 120(4): p. 1140–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Allen DG, Zhang BT, and Whitehead NP, Stretch-induced membrane damage in muscle: comparison of wild-type and mdx mice. Adv Exp Med Biol, 2010. 682: p. 297–313. [DOI] [PubMed] [Google Scholar]
- 99.Childers MK, et al. , Eccentric contraction injury in dystrophic canine muscle. Arch Phys Med Rehabil, 2002. 83(11): p. 1572–8. [DOI] [PubMed] [Google Scholar]
- 100.Childers MK, et al. , Myofiber injury and regeneration in a canine homologue of Duchenne muscular dystrophy. Am J Phys Med Rehabil, 2001. 80(3): p. 175–81. [DOI] [PubMed] [Google Scholar]
- 101.Childers MK, et al. , Skinned single fibers from normal and dystrophin-deficient dogs incur comparable stretch-induced force deficits. Muscle Nerve, 2005. 31(6): p. 768–71. [DOI] [PubMed] [Google Scholar]
- 102.De Pooter J, et al. , Elevated troponin T levels in a female carrier of Duchenne muscular dystrophy with normal coronary angiogram: a case report and review of the literature. Acta Cardiol, 2012. 67(2): p. 253–6. [DOI] [PubMed] [Google Scholar]
- 103.Deconinck N. and Dan B, Pathophysiology of duchenne muscular dystrophy: current hypotheses. Pediatr Neurol, 2007. 36(1): p. 1–7. [DOI] [PubMed] [Google Scholar]
- 104.Grounds MD, Two-tiered hypotheses for Duchenne muscular dystrophy. Cell Mol Life Sci, 2008. 65(11): p. 1621–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Phillips MF and Quinlivan R, Calcium antagonists for Duchenne muscular dystrophy. Cochrane Database Syst Rev, 2008(4): p. CD004571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Perel P, et al. , Comparison of treatment effects between animal experiments and clinical trials: systematic review. BMJ, 2007. 334(7586): p. 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dimos JT, et al. , Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science, 2008. 321(5893): p. 1218–21. [DOI] [PubMed] [Google Scholar]
- 108.Lee G, et al. , Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature, 2009. 461(7262): p. 402–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Marchetto MC, et al. , A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell, 2010. 143(4): p. 527–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Farra N, et al. , Rett syndrome induced pluripotent stem cell-derived neurons reveal novel neurophysiological alterations. Mol Psychiatry, 2012. 17(12): p. 1261–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Brennand KJ, et al. , Modelling schizophrenia using human induced pluripotent stem cells. Nature, 2011. 473(7346): p. 221–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wernig M, et al. , Neurons derived from reprogrammed fibroblasts functionally integrate into the fetal brain and improve symptoms of rats with Parkinson’s disease. Proc Natl Acad Sci U S A, 2008. 105(15): p. 5856–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pasca SP, et al. , Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat Med, 2011. 17(12): p. 1657–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yazawa M, et al. , Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature, 2011. 471(7337): p. 230–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Salva MZ, et al. , Design of tissue-specific regulatory cassettes for high-level rAAV-mediated expression in skeletal and cardiac muscle. Mol Ther, 2007. 15(2): p. 320–9. [DOI] [PubMed] [Google Scholar]
- 116.Pharmaceutical Research and Manufacturers of America Profile, 2009, PhRMA: Washington, DC. [Google Scholar]
- 117.Hughes B, 2009 FDA drug approvals. Nat Rev Drug Discov, 2010. 9(2): p. 89–92. [DOI] [PubMed] [Google Scholar]
- 118.Holland J. Fixing a broken drug development process. Journal of Commercial Biotechnology, 2013. 19, 5–6. [Google Scholar]
- 119.FitzGerald GA, Re-engineering drug discovery and development. LDI Issue Brief, 2011. 17(2): p. 1–4. [PubMed] [Google Scholar]
- 120.Zhou H, et al. , Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell, 2009. 4(5): p. 381–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kim D, et al. , Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell, 2009. 4(6): p. 472–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Warren L, et al. , Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell, 2010. 7(5): p. 618–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Obokata H, et al. , Stimulus-triggered fate conversion of somatic cells into pluripotency. Nature, 2014. 505(7485): p. 641–7. [DOI] [PubMed] [Google Scholar]