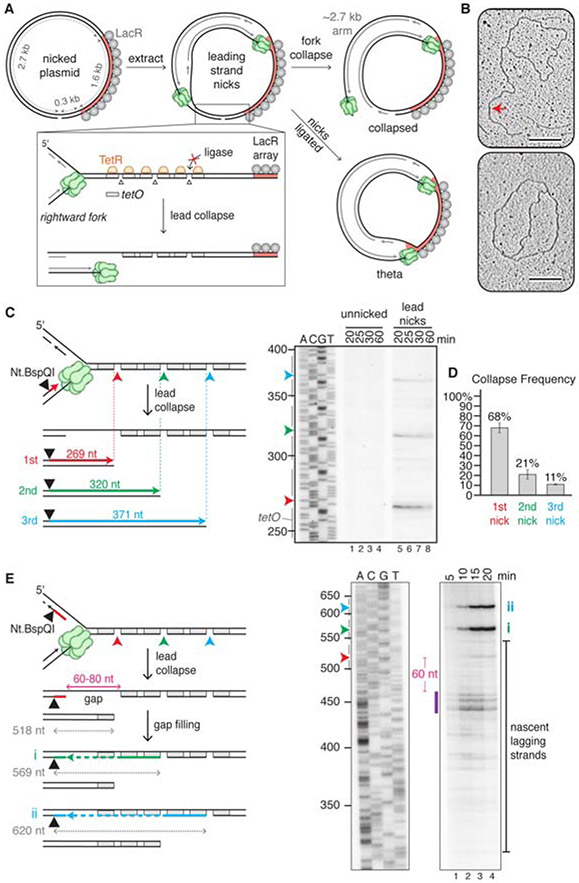
Figure 1. Lead collapse generates a nearly blunt seDSB and a gap in the lagging strand.
(A) Experimental approach used in Figures 1 and 3. (B) Plasmid nicked with Nb.BbvCI was incubated with TetR and LacR and replicated in egg extract for 15 minutes before extracting the DNA for electron microscopy. Collapsed (top) and theta (bottom) structures are shown. Red arrow, 2.7 kb arm. Scale bar, 200 nm. Images are representative from two biological replicates. (C) Analysis of nascent leading strands. Nb.BbvCI-nicked plasmid was replicated as in panel B, but in the presence of [a-32P]dATP. After isolation, DNA was digested with Nt.BspQI, separated via denaturing electrophoresis, and subjected to autoradiography. Only the relevant segment of the gel is shown. Gray bars, tetO sites. (D) Percentage collapse at each of the three nicks in panel C. Error bars, standard deviations from three biological replicates. (E) Repetition of panel C, but with the Nt.BspQI site located on the nascent lagging strand. After fork collapse at the first nick, gap filling from the 3’ end of the nick (still protected by TetR) to the final Okazaki fragment and ligation creates a 569 nt product (i, green line and arrowhead). Collapse at the second nick, gap filling, and ligation generates a 620 nt product (ii, blue line and arrowhead). Purple bar, most prominent lagging strand products. Irrelevant lanes between the two panels shown were removed. Gels are representative images from three biological replicates. See also Figure S1.