Summary
The high-throughput phenotypic screen (HTPS) has become an emerging technology to discover synthetic small molecules that regulate stem cell fates. Here, we review the application of HTPS to identify small molecules controlling stem cell renewal, reprogramming, differentiation, and lineage conversion. Moreover, we discuss the use of HTPS to discover small molecules/polymers mimicking the stem cell extra-cellular niche. Furthermore, HTPSs have been applied on whole animal models to identify small molecules regulating stem cell renewal or differentiation in vivo. Finally, we discuss the examples of the utilization of HTPS in both stem cell-based disease modeling, as well as the discovery of novel drug candidates for cancer, diabetes, and infectious diseases. Overall, HTPSs have provided many powerful tools for the stem cell field, which not only facilitate the generation of functional cells/tissues for replacement therapy, disease modeling, and drug screening, but also help dissect molecular mechanisms regulating physiological and pathological processes.
Keywords: Stem cell fate, Stem cell niche, Target identification, Disease Modeling, Drug screening
Graphical Abstract
eTOC blurb
Vandana et al. highlight the role of HTPS as a powerful tool in stem cell biology to unearth small molecules that control cell specification and the stem cell niche, expanding the repository of novel candidates available for use in disease modeling or therapy.
Introduction
Stem cells have the potential to develop into many different types of cells in the body. Stem cells are defined by two characteristics: self-renewal, or the ability to proliferate and replenish themselves over time, and differentiation, or the ability to form committed progenitors and, ultimately, terminally differentiated cells. Stem cells are classified into two categories: pluripotent stem cells (PSCs), including embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs), and somatic stem cells (SSCs). Mouse ESCs (mESCs) were first isolated from the inner cell mass (ICM) of early embryos or blastocysts (Evans and Kaufman, 1981; Martin, 1981). Human ESCs (hESCs) were isolated from blastocysts in 1998 (Thomson et al., 1998). A revolutionary advancement was further made in 2006 and 2007, when iPSCs were derived via the reprogramming of mouse and/or human fibroblasts by four defined reprogramming factors: Oct3/4, Sox2, Klf4, and c-Myc (OSKM) or Oct3/4, Sox2, Nanog, and Lin-28 (Takahashi et al., 2007; Takahashi and Yamanaka, 2006; Yu et al., 2007). SSCs, such as hematopoietic stem cells, mesenchymal stem cells, and intestinal stem cells, have been identified in and isolated from multiple organs (Jung et al., 2011; Pittenger et al., 1999). These SSCs play critical roles in tissue homeostasis and regeneration.
One key question in the field of stem cell biology is how to control stem cell fate decisions, such as self-renewal, differentiation, and reprogramming. Embryonic development comprises of a sequence of highly concerted events, including the expression of specific genes, soluble factors, and other proteins at appropriate levels with precise spatiotemporal regulation. By utilizing the chemicals and growth factors mimicking the developmental processes in vivo, significant progress has been made to control stem cell fate decision at different stages. For example, retinoic acid, which establishes the anterior-posterior axis in vertebrate organogenesis, is critical for cell differentiation and commonly incorporated during stem cell differentiation in vitro (Cunningham and Duester, 2015). However, endogenous small molecules or growth factors that regulate cell fate are limited. The lack of efficient differentiation protocols to derive mature cells prompts the need to screen for novel chemical compounds beyond those that make up the developmental toolbox at present.
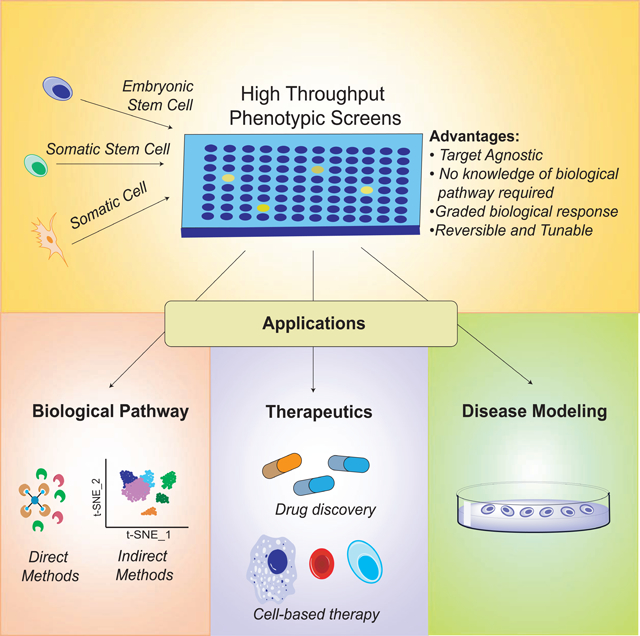
Phenotype-based approaches possess several key advantages over traditional target-based approaches in that they are target-agnostic and one does not need to possess specific knowledge pertaining to cellular pathways (Ursu et al., 2017). As synthetic or in silico approaches increase the accessibility to a vast chemical space, the HTPS approach serves as an attractive option towards deconvoluting complex biological processes (Hoffmann and Gastreich, 2019). Specifically, chemical compounds provide insights on graded biological responses, as they are able to reversibly and selectively tune biological pathways. This paves the way for the development of mature, functional cells/tissues/organs for regenerative medicine (Figure 1).
Figure 1. Schematic of HTPSs to identify synthetic small molecules controlling stem cell fate decision.

Different cell types are treated with compounds from chemical libraries following which one of the detection methods including immunocytochemistry-based, morphology-based, reporter-based, or cell viability-based methods are used. Following the induction of the desired phenotype, compounds that regulate pluripotency, reprogramming, differentiation, lineage reprogramming in different cell types can be identified and confirmed.
To identify regulators of biological pathways, HTPSs are typically performed using chemically diverse small molecule libraries using appropriate readouts. Some of the most common readouts are based on cell viability, reporter activity, immunocytochemistry or morphology (Figure 1). Cell viability assays are typically able to distinguish between mitochondrial activity, cell metabolism and/or enzyme activity of live and dead cells and are suitable for the discovery of compounds that improve PSC or SSC survival. Reporter assays and immunocytochemistry based assays can be indicative of a specific signaling pathway. Reporter based assays are particularly efficient for rapid discovery in HTPSs because of the direct measure of reporter activity by detection of a luminescent or fluorescent signal while immunocytochemistry based screens are more labor intensive and subject to variation. However, it is important to note that it is necessary to establish stable cell lines expressing the appropriate reporter gene in order to adopt this assay. Phenotypic changes pertaining to disease states can additionally be evaluated by morphological changes or immunocytochemistry. This has been applied to drug discovery using isogenic PSCs containing disease-associated mutations/variants which recapitulate dysregulated cellular functions.
In this review, we will elaborate on the application of HTPSs to identify chemical candidates or biomaterials that control stem cell fate decision. Moreover, we will discuss recent studies utilizing HTPSs for stem cell-based drug discovery. Finally, we will highlight recent technologies that have emerged to explore prospective targets or mechanisms of action following the identification of hit compounds in HTPSs.
Chemical modulators of pluripotency
Since the isolation of the first ESC line, HTPSs have been applied to identify synthetic small molecules to maintain the pluripotency of ESCs, which facilitates the transition of ESC culture to chemically-defined feeder-free systems. First, a HTPS used Oct4-GFP mESCs to identify, SC1, which maintains mESCs pluripotency by dual inhibition of Ras GTPase-activating protein (RasGAP) and extracellular-signal-regulated kinase I (ERK1) (Chen et al., 2006). In addition, a HTPS monitoring the alkaline phosphatase (ALP) activity identified IQ-1, which maintains the stemness of mESCs in the absence of leukemia inhibitory factor (LIF) by upregulating ß-catenin/CBP-mediated transcription (Miyabayashi et al., 2007). Nanog is another integral protein required for the maintenance of the undifferentiated state. A HTPS utilizing Nanog-GFP reporter mESCs screened 18,360 compounds from the RIKEN natural products depository library (NPDepo) and identified NPD13432, a GSK-3 inhibitor, to maintain mESC self-renewal (Kobayashi et al., 2020). Adapting hPSCs to a HTPS format revealed several candidates including antibiotics, anti-inflammatory drugs, and cardiac glycosides that promote self-renewal or differentiation of hPSCs upon screening a library of 2,880 small molecules and detecting OCT4 expression via high-content imaging (Desbordes et al., 2008).
The poor survival of PSCs has also prompted the screen for compounds that increase overall survival. Regulators of hESC cell survival have been discovered including highly specific ROCK inhibitors and PKA/C inhibitors using HTPS (Damoiseaux et al., 2009). Automated multi-parametric analysis including cell count per colony, colony count, nuclear staining, TRA-1-60 staining, and morphology has been incorporated in a HTPS study, revealing pinacidil and several steroids as potent regulators of stem cell survival and differentiation respectively (Barbaric et al., 2010).
Although PSCs lie at the top of the hierarchy in terms of developmental potential, they lack the ability to form extraembryonic tissues. Recently, by activating the Oct4 distal enhancer (Oct4-DE) and eliminating transforming growth factor (TGF)-β signaling dependency, a minimal chemical cocktail of hLIF, CHIR 99021, (S)-(+)-dimethindene maleate, and minocycline hydrochloride, has been identified to induce the formation of extended PSCs (EPS) with bi-potential toward both lineages (Yang et al., 2017).
Chemical modulators of reprogramming
Immediately after the discovery of OSKM-mediated reprogramming, significant efforts were applied to discover synthetic small molecules that could potentially replace one or more transcription factors. Many HTPSs of reprogramming identified epigenetic modulators, which play a role in altering the cellular methylation or acetylation landscape (Table 1). First, a HTPS based on ALP activity identified DNA methyltransferase (DNMT) inhibitors, including RG108 and 5-azacytidine, which synergize with BIX-01294, a G9a methyltransferase inhibitor, to reprogram MEFs into mouse iPSCs (miPSCs) in the absence of Sox2 (Shi et al., 2008). Valproic acid (VPA) was previously reported as a HDAC inhibitor, capable of enhancing reprogramming efficiency in MEFs, hemizygous for Oct4-GFP, in the absence of c-myc or Klf4 with only the genetic factors, Oct4 and Sox2 (Huangfu et al., 2008a; Huangfu et al., 2008b). A HTPS using a fluorescent reporter, Cdy1, led to the discovery of another HDAC inhibitor, 1–26 (Vendrell et al., 2012), which promotes reprogramming in the absence of c-myc. Recently, Headley et al. identified mocetinostat as a potent inducer of reprogramming via epigenetic alterations of the Oct4 locus using a chromatin in vivo mouse platform (Headley et al., 2019). Furthermore, a chromatin focused screen identified the acetyl-lysine competitive inhibitors, CBP30 and I-CBP112, which accelerate reprogramming to hiPSCs using only OCT4 and SOX2 in combination with DOT1L inhibition (Ebrahimi et al., 2019).
Table 1.
HTPSs to identify chemical modulators of reprogramming
| Target | Starting population | Library | Screening Format | Readout | Hit Compounds | Mechanism of Action | Reference |
|---|---|---|---|---|---|---|---|
| Replacing Sox2 | MEFs | 2,000 known bioactive molecules | 6-well plate | ALP activity and morphology | BIX-01294 and BayK8644 | Inhibition of G9a mediated H3K9me2 and potential effect on calcium signaling | (Shi et al., 2008) |
| Oct4-GFP MEFs | 200 compounds from a library of compounds with known targets | 96-well plate | GFP expression | SB431542 and RepSox | Inhibition of TGF-β signaling | (Ichida et al., 2009) | |
| Nanog-Luc MEFs | Collection of 750,000 chemically diverse compounds | 1536-well plate | Luciferase activity | iPyrazine | Inhibition of pan-Src family kinases | (Staerk et al., 2011) | |
| Replacing cMyc | MEFs | Collection of 240 synthesized hydroxamic acid derivatives | 384-well plate | CDy1 fluorescence | 1–26 | Inhibition of HDACs | (Vendrell et al., 2012) |
| Replacing Klf4 | Nanog-Luc MEFs | Collection of 500,000 chemically diverse compounds | 1536-well plate | Luciferase activity | Kenpaullone | GSK3β inhibitor, CDK inhibitor | (Lyssiotis et al., 2009) |
| Overcoming epigenetic barriers | CiA-Oct4 MEFs | 959 molecules from an epigenetic-targeted library | 384-well plate | GFP expression | Mocetinostat | Inhibition of HDACs | (Headley et al., 2019) |
| Human fibroblasts | 60 compounds | 96-well plate | TRA-1-60 staining | CBP30 and I-CBP112 | Competitive inhibition of acetyl-lysines | (Ebrahimi et al., 2019) | |
| Oct4-GFP MEFs | Collection of 100 compounds | 12-well plate | GFP expression | Combination of AMI-5 and A-83-01 | Inhibition of PRMT 1/3/4/6 | (Yuan et al., 2011) | |
| Improving reprogramming efficiency | Neonatal human epidermal keratinocytes | Collection of more than 50 known bioactive compounds | 10-cm plates | TRA-1–81 staining | PS48, fructose-2,6-bisphosphate, oxalate, 2,4-dinitrophenol, N-oxaloylglycine, and quercetin | Stimulation of glycolytic metabolism | (Zhu et al., 2010) |
| Nanog-GFP MEFs | More than 1600 known bioactive compounds | 384-wellplate | GFP expression | Rapamycin and AICAR | AMPK-dependent autophagy and mitochondrial clearance | (Ma et al., 2015) | |
| MEFs | 665 FDA-approved compounds | 96-well plate | ALP staining | Fenofibrate | Reducing oxidative stress by PPARα stimulation | (Lee et al., 2018) | |
| Replacing OSKM with small molecules | Oct4-GFP MEFs | 10,000 compounds from various libraries | 12-well plate | GFP expression and morphology analysis | VPA, CHIR99021, 616452, tranylcypromi ne, FSK, and DZNep | (Hou et al., 2013) |
Another group of compounds promoting reprogramming include modulators of endogenous signaling processes. A HTPS based on Oct4-GFP MEFs identified RepSox and SB431542, the TGF-β pathway regulators, which facilitate reprogramming in the absence of Sox2 (Ichida et al., 2009). Using a HTPS based on a Nanog-Luciferase (Luc) MEFs, Lyssiotis et al screened 500,000 compounds and reported, kenpaullone, a potent GSK3 inhibitor, which promotes reprogramming with a subset of reprogramming factors lacking Klf4 (Lyssiotis et al., 2009). Moreover, Yuan et al used a synergistic chemical screen based on Oct4-EGFP MEFs and identified AMI-5, a protein arginine methyltransferase inhibitor, which increases the efficiency of Oct4-induced reprogramming of MEFs in combination with a TGF-β inhibitor, A-83-01 (Yuan et al., 2011). Furthermore, Staerk et al performed a HTPS using Nanog-Luc MEFs and identified iPYrazine, which replaces Sox2 during reprogramming by inhibiting Pan-Src family kinase (Staerk et al., 2011).
Recently, several HTPSs identified metabolic regulators that facilitate reprogramming. Using a HTPS based on the number of TRA-1-81+ colonies, Zhu et al screened a collection of known bioactive compounds and found PS48, a pyruvate dehydrogenase kinase isozyme 1 activator, stimulates human fibroblast reprogramming with OCT4. They also found that other compounds that promote glycolysis, such as fructose-2,6-bisphosphate, oxalate, 2,4-dinitrophenol, N-oxaloylglycine, and quercetin, show similar abilities to facilitate reprogramming (Zhu et al., 2010). Using a HTPS by measuring Nanog-GFP reporter signal and colony formation, Ma et al screened over 1,600 compounds and identified rapamycin and AICAR, which increase the reprogramming efficiency through mediating AMPK-dependent mitochondrial clearance (Ma et al., 2015). Recently, Lee et al performed a HTPS based on ALP+ colonies and identified PPARα agonists, such as fenofibrate, increase the colony survival rate during reprogramming by mediating PPARα stimulation of antioxidant and detoxifying enzymes (Lee et al., 2018).
A remarkable advancement in iPSC reprogramming occurred with the discovery that a cocktail of seven small molecules could form chemically induced PSCs (ciPSCs) from MEFs (Hou et al., 2013; Long et al., 2015). A later study revealed that it was also possible to derive ciPSCs from mouse neural stem cells and small intestinal epithelial cells (Ye et al., 2016).
HTPS to Identify Chemical Modulators of Stem Cell Differentiation
Cell-based therapy and disease modeling in a dish face exciting prospects, especially with the directed differentiation of PSCs along specific lineages. Many HTPSs were performed to identify chemical compounds that direct PSCs differentiation, particularly pertaining to cardiomyocytes, neurons, and beta cells (Figure 2 and Table 2).
Figure 2. Representative structures of chemical candidates for directed differentiation.
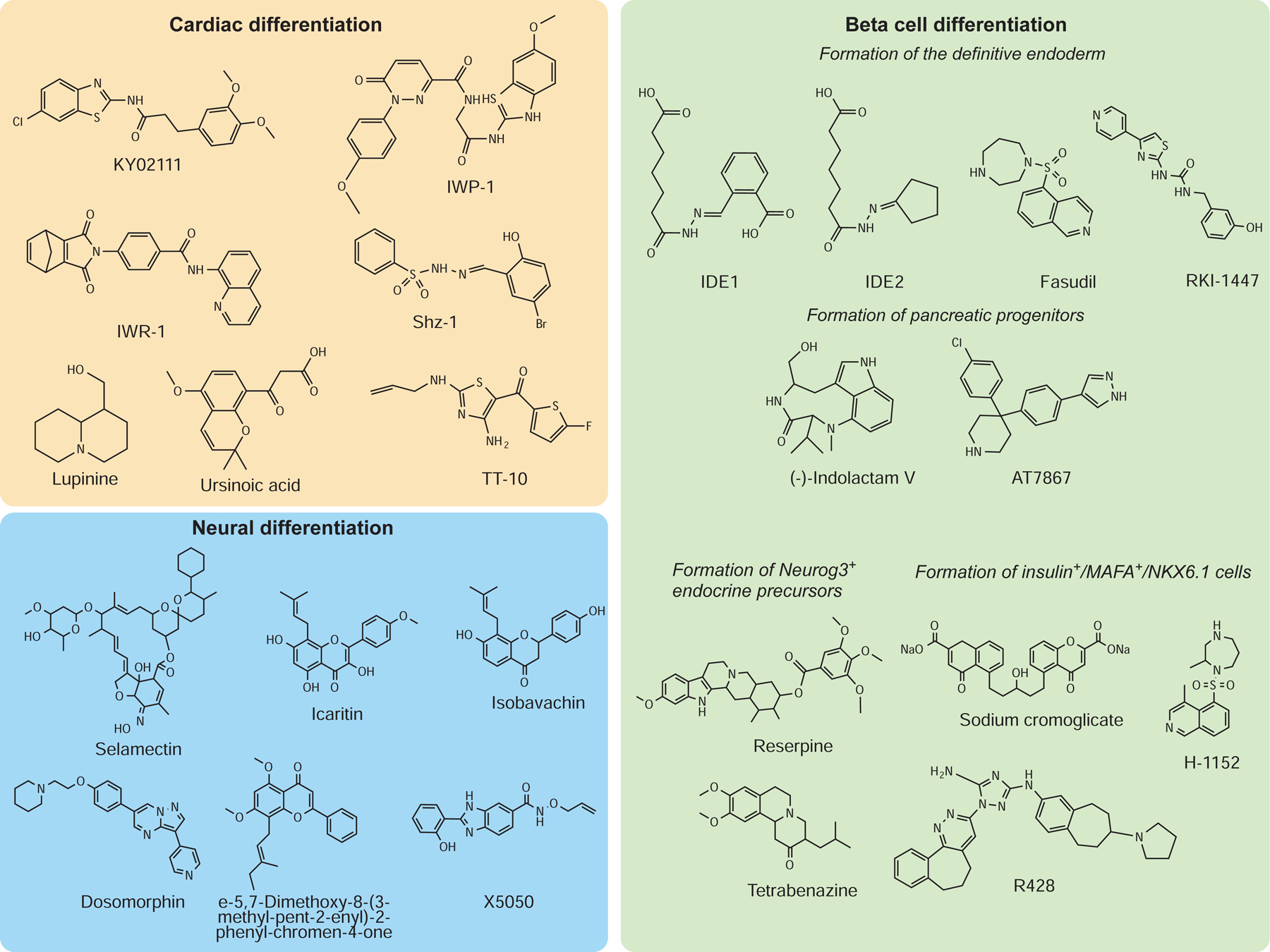
Cell types of the mesodermal, ectodermal, and endodermal lineage have been obtained by the use of chemical modulators that promote the conversion of PSCs into cardiac, neuronal, and beta cells.
Table 2.
HTPSs to identify chemical modulators of directed differentiation
| Target | Starting Population | Library | Screening Format | Readout | Hit Compounds | Mechanism of Action | Reference |
|---|---|---|---|---|---|---|---|
| Cardiac differentiation | MHC-GFP mESCs | 880 FDA approved drugs | 96-well plate | GFP expression | Ascorbic acid | Modulation of SMAD signaling | (Takahashi et al., 2003) |
| ANF-Luc mESCs | Collection of 100,000 heterocyclic compounds | 384-well plate | Luciferase activity | Cardiogenol (A-D) | (Wu et al., 2004) | ||
| Nkx2.5-Luc mESCs | 47,000 compounds from DiverSet library and 100,000 compounds from Chemical diversity Labs | 96-well plate | Luciferase activity | Sulfonylhydrazone (shz) molecules | (Sadek et al., 2008) | ||
| MHC-GFP monkey ESCs | Collection of 9,600 compounds | 96-well plate | GFP expression | KY02111 | Inhibition of WNT signaling | (Minami et al., 2012) | |
| MYH6-mCherry hESCs | 550 known pathway modulators; 244 kinase inhibitors and 305 pathway agonists and antagonists | 384-well plate | mCherry expression | IWR-1 and IWP | Inhibition of WNT signaling | (Willems et al., 2011) | |
| MHC-mCherry mESCs | 800 compounds comprising of diverse alkaloids, flavonoids and sterols | 96-well plate | mCherry expression | Lupinine and ursinoic acid | Early induction of WNT signaling | (Lee et al., 2019) | |
| CCS:Lacz ESCs | 1,280 pharmacologi cally active compounds, 2,320 compounds and 1,200 compounds including 100% FDA-approved drugs | 384-well plate | β-galactosidase activity | Sodium nitroprusside | Activation of cAMP signaling | (Tsai et al., 2015) | |
| Neuronal differentiation | mESCs | 2,000 compounds comprising of FDA-approved drugs, bioactive compounds, and natural products | 96-well plate | Tyrosine hydroxylase (TH) staining | Selamectin | Targets the γ2 subunit-containing GABAA receptor | (Sun et al, 2013) |
| REST-Luc NSCs | 5,864 compounds from the CHEM-X Infinity library and 1,120 compounds from the Prestwick library | 384-well plate | Luciferase activity | X5050 | (Charbord et al.,2013) | ||
| Beta cell differentiation | Sox17-dsRed mESCs | 2,000 bioactive compounds and known drugs, 1,000 synthetic HDAC inhibitors, 20 modulators of stem cell fate, 400 compounds including bioactives and natural products | 384-well plate | dsRed expression | IDE1 and IDE2 | Activation of TGF-β signaling | (Borowiak et al., 2009) |
| mESCs | 23,406 compounds from the Enamine screening set with drug-like properties and FDA-approved compounds | 384-well plate | FoxA2 staining | Fasudil and RKI-1447 | ROCK inhibition | (Korostylev et al., 2017) | |
| Endoderm derived from hESCs | 5,000 compounds including pathway regulators and kinase inhibitors | 384-well plate | Pdx-1 staining | (−)-Indolactam V | PKC signaling | (Chen et al., 2009) | |
| PDX1+PPs derived from hiPSCs | 1,327 kinase inhibitors | 96-well plate | Cell number | AT7867 | (Kimura et al., 2017) | ||
| INS+PDX+ cells derived from hESCs | FDA approved drug library, ICCB Bioactive Library, US and international Drug Collection, LOPAC1280, and Prestwick Library | 384-well plate | Insulin staining | Sodium cromoglicate | Modulation of chloride channel, HSP 90, or GPCR 35 signaling | (Kondo et al., 2017) | |
| INS+PDX+ cells derived from hESCs | Over 40 different compounds | MAFA gene expression | R428 | Inhibition of AXL | (Rezania et al., 2014) | ||
| PDX1+PPs derived from hiPSCs | More than 4000 compounds consisting of FDA approved drugs, kinase inhibitors, and signaling pathway regulators | 384-well plate | Insulin staining | H1152 | ROCKII inhibition | (Ghazizadeh et al., 2017) | |
| PPs derived from hiPSCs | 1,120 biologically active compounds derived from the Prestwick library | 96-well plate | Insulin staining | Reserpine, tetrabenazine | VMAT2 inhibition | (Sakano et al., 2014) | |
| Mesoderm differentiation | OSR1-GFP hiPSCs | 1,800 compounds from the Prestwick and ENZO library | 96-well plate | GFP expression | AM580 and TTNPB | Retinoic acid signaling | (Araoka et al., 2014) |
| Myotube maturation | Myogenic progenitors derived from hESCs/hiPSCs | 80 compounds from the Tocris library | 96-well plate | MHC staining | SB431542, DAPT, Dexamethasone, Forskolin, PD032590 | Inhibition of TGF-β and Notch signaling | (Selvaraj et al., 2019) |
The regenerative capacity of adult cardiomyocytes is limited and, hence, there is an imminent need for cell-based therapy for cardiomyopathies. Using cardiac-specific α-cardiac myosin heavy chain (Mhc) promoter-driven EGFP, Takahashi et al screened 880 compounds approved for human use and identified ascorbic acid as an inducer of cardiac differentiation in mESCs (Takahashi et al., 2003). Wu et al used a mouse embryonic carcinoma (EC) cell line, P19, carrying rat atrial natriuretic factor (Anf) and identified that cardiogenol C promotes cardiogenesis (Wu et al., 2004). Using α-MHC promoter-driven EGFP transgenic monkey ESCs, Minami et al identified KY02111 as a cardiac differentiation inducer that inhibits the WNT pathway (Minami et al., 2012). Moreover, IWR-1 and IWP, other WNT signaling inhibitors, have been reported to produce beating foci from monolayer cultures (Willems et al., 2011). Recently, Lee et al identified the natural alkaloids, lupinine and ursinoic acid, as stimulators of early WNT signaling, which promotes cardiac mesoderm generation (Lee et al., 2019). By screening a chemical library for activators of the signature gene Nkx2.5, Sadek et al identified sulfonylhydrazone (Shz) molecules trigger myocardin, troponin I and sarcomeric α-tropomyosin expression (Sadek et al., 2008). Using a HTPS based on CCS:lacz mESCs, Tsai et al discovered sodium nitroprusside, which promotes the generation of cardiac Purkinje cells by activating cyclic AMP signaling (Tsai et al., 2015). Ito et al, reported a compound, TT-10, that stimulates the cell cycle of hiPSC-derived cardiomyocytes by targeting the YES-associated protein (YAP)/transcriptional enhancer factor domain (TEAD) axis using HTPS (Ito et al., 2019).
In addition to cardiac differentiation, another key area of interest for directed differentiation screens is neuronal lineages. Early studies identified cellular signaling modulators, such as icaritin and isobavachin, which stimulate neuronal differentiation via the simultaneous inhibition and activation of the p38-MAPK and ERK pathways, respectively (Wang et al., 2011; Wang et al., 2009). Mei et al reported the discovery of a flavonoid, 5,7-dimethoxy-8-(3-methyl-pent-2-enyl)-2-phenyl-chromen-4-one, which was found to exert its effect on neuronal differentiation by activating PPARß and, consequently, tuning mitochondrial metabolism through MFN2 expression and mitochondrial Ca2+ levels (Mei et al., 2016). In addition, Zhou et al reported that dorsomorphin plays a role in the selective conversion of PSCs into neural cells by inhibiting BMP and Activin signaling (Zhou et al., 2010). A combinatorial screen further identified chemical inhibitors, including SU5402, CHIR99021, and DAPT, which promote the rapid conversion of hPSCs into P2RX3 expressing nociceptors when administered in combination with LDN-193189 and SB431542 (Chambers et al., 2012). Using a HTPS of ~2000 bioactive compounds in mESC monolayer cultures labeled with the anti-tyrosine hydroxylase (TH) antibody (a marker for dopaminergic, noradrenergic, and adrenergic neurons), Sun et al found that selamectin could bind to γ-containing GABAA receptors in neural rosette progenitors, promoting the expression of neural transcription factors (Sun et al., 2013). Finally, a luciferase based HTPS measuring REST activity in neural derivatives of hESCs led to the identification of a benzoimidazole-5-carboxamide derivative, X5050, which increased the expression of neuronal genes such as BDNF and SNAP25 (Charbord et al., 2013).
Finally, HTPS have also been applied to facilitate the discovery of modulators that commit PSCs to endodermal lineages. For instance, a flow cytometry based-screen using mESCs carrying a Sox17-dsRed reporter identified IDE1 and IDE2, which synergize with low doses of Activin A or Nodal to commit mESCs and hESCs to the endodermal lineage (Borowiak et al., 2009). Using a HTPS based on FOXA2 expression, Korostylev et al identified Fasudil and RKI-1447, two potent ROCK inhibitors, direct the differentiation of mESCs to form the definitive endoderm (Korostylev et al., 2017). Following the establishment of the definitive endoderm, PDX+ pancreatic progenitor (PP) cell formation is necessary to generate functional beta cells. At this step, (−)-indolactam V and AT7867 were discovered as robust inducers of PP formation (Chen et al., 2009; Kimura et al., 2017). In the later stages of differentiation, sodium cromoglicate has been found to stimulate the formation of insulin+ cells from PDX+ PP cells (Kondo et al., 2017). However, these cells do not achieve complete maturation, in which several key markers, including MAFA and NKX6.1, are absent. In contrast, a small scale screening identified R428, an inhibitor of the tyrosine kinase receptor AXL, as an inducer of MAFA, which gives rise to insulin+/glucagon−/somatostatin+ cells in combination with ALK5 inhibitor and T3 (Rezania et al., 2014). A HTPS using an insulin antibody identified vascular monoamine transporter 2 (VMAT2) inhibitors, such as reserpine and tetrabenazine (TBZ), which act synergistically with dibutyryl adenosine 3’, 5’-cyclic AMP to promote differentiation towards Neuroginin-3+ endocrine precursors (Sakano et al., 2014). Finally, another HTPS demonstrated that ROCKII inhibition plays a role in beta cell maturation. H1152 was able to increase the percentage of insulin+ cells and glucose stimulated insulin secretion (GSIS) (Ghazizadeh et al., 2017).
In addition to the three lineages as described above, HTPS has been applied to screen for compounds that promote hepatogenesis, nephrogenesis, formation of intestinal cells, and the formation of myotubes (Araoka et al., 2014; Kabeya et al., 2018; Selvaraj et al., 2019; Shan et al., 2013). Overall, the discovery of a myriad of chemical candidates that promote lineage-specific differentiation in a stage-specific manner has significantly enhanced the efficiency and maturation status of hPSC-derived cells/tissues.
Chemical modulators of lineage reprogramming
Lineage reprogramming, circumventing the pluripotent state, serves as a quicker means of attaining the desired cell type. Most lineage reprogramming processes require a combination of chemical compounds targeting multiple signaling pathways. A HTPS using a collection of 500,000 compounds identified a chemical cocktail including forskolin, ISX9, CHIR99021, and I-BET151 or SB431542, promotes the reprogramming from fibroblasts into TAU-EGFP+/TUJ1+ positive neuronal cells (Li et al., 2015b). Moreover, the conversion of human astrocytes into neurons was achieved by a combination of 9 master conversion molecules, including LDN193189, SB431542, TTNPB, Tzv, CHIR99021, VPA, DAPT, SAG, and Purmo, added in a stepwise manner (Zhang et al., 2015). A study incorporating cell activation and signaling directed (CASD) lineage conversion determined A83-01, CHIR99021, sodium butyrate, LPA, rolipram, and SP600125 as necessary components for OCT4-mediated conversion of fibroblasts into human neural stem cells (Zhu et al., 2014). The successful derivation of MAP2+ neurons from fibroblasts was further achieved using a cocktail comprising of PGE2, forskolin, BML210, amino resveratrol sulfate, and PP2, which were identified from a HTPS comprised of compounds from five annotated libraries (Pfisterer et al., 2016). In addition, Mohamed et al. screened 5,500 compounds and reported a combination of the TGF-β inhibitor, SB431542, and the WNT inhibitor, XAV939, reprograms cardiac fibroblasts to cardiomyocyte-like cells both in vitro and in vivo (Mohamed et al., 2017). Lumelsky reported a combination of retinoic acid, A83-01, pVc, LDE225, and Bix-01294 converted fibroblasts to islet-like cells (Lumelsky, 2014). A combinatorial screening revealed the compounds valproic acid, CHIR98014, Repsox, TTNPB, and celecoxib, were essential for the transdifferentiation of MEFs into fibrocartilaginous cells (Chen et al., 2020). In summary, HTPSs have identified several chemical combinations for lineage reprogramming.
Chemical modulators of SSCs
Since SSCs are limited in terms of their proliferation capacity, HTPSs have been widely adopted to identify compounds that promote SSC expansion. Prostaglandin E2 (PGE2) was found to boost HSC formation by activating ß-catenin via cAMP-mediated activation of PKA (North et al., 2007). The discovery of PGE2 then led to the development of the long-acting derivative 16,16-dimethyl-PGE2 (dmPGE2) that accelerates engraftment in human umbilical cord blood (UCB) transplantation in Phase I clinical trials (Hagedorn et al., 2014). Compounds that modulate blood flow, including agents that act on α- and β-adrenergic receptors, calcium flux modulators, NO signaling modulators, and angiotensin converting enzyme (ACE) regulators were revealed to enhance runx1/cmyb+ HSC formation (North et al., 2009). StemReginin 1 (SR1) and UM171 were identified to upregulate the HSC expansion via aryl hydrocarbon receptor dependent and independent pathways respectively (Boitano, 2011; Fares et al., 2014). VPA was found to increase the number of CD34+ cells that are derived from both peripheral and cord blood stem cells (Arulmozhivarman et al., 2017). Moreover, Li et al, identified epoxyeicosatrienoic acids improve the homing and engraftment efficiency of HSCs from a HTPS using the ICCB Known Bioactive library (Li et al., 2015a). In addition, a bioluminescence imaging based HTPS revealed ergosterol as a candidate that improves HSC homing in zebrafish and mice by increasing CXCR4 expression (Astuti et al., 2017).
Several HTPSs have been applied to study mesenchymal stem cell (MSC) differentiation. A HTPS based on a glycosaminoglycan assay (GAG) revealed several potent inducers and inhibitors of chondrogenesis including hypnotic, anti-neoplastic, and anti-protein synthesis compounds (Huang et al., 2008). An epigenetic library screen identified abexinostat as a regulator of hMSC differentiation into both osteoblasts and adipocytes (Ali et al., 2016). In a recent study detecting ALP activity levels, DIPQUO was identified as an activator of the p38-MAPK pathway, increasing the expression of RUNX2, Osterix, and Osteocalcin in both human MSCs and in zebrafish (Cook et al., 2019). Another reporter-based assay has revealed compounds, such as E05657 and E09241, which increase the OPG/RANKL ratio and OPG secretion, decrease NFATC1 expression, and reduce osteoclastogenesis (Gong et al., 2016; Han et al., 2019). The identification of numerous compounds that promote osteogenesis could facilitate in vivo tissue regeneration.
Screening for extracellular matrix for stem cell culture
The extracellular matrix (ECM) plays a critical role in controlling stem cell fate decisions. Several HTPSs have been applied to develop appropriate extracellular matrices for different stem cell cultures. For example, a HTPS study explored the extracellular proteome as a source of effector proteins to regulate stem cell fate by screening 806 purified secreted proteins, which led to the discovery of the pigment epithelium-derived factor as a modulator of hESC self-renewal (Gonzalez et al., 2010). Brafman et al reported a combinatorial screen of 7 extracellular matrix proteins which led to the identification of fibronectin and vitronectin that promotes endodermal differentiation (Brafman et al., 2013). Moreover, peptides such as the cell adhesion ligand, GRGDSP, have been found to stimulate ALP activity in hMSCs (Koepsel et al., 2012).
Interestingly, synthetic materials devoid of extracellular matrix proteins have also been evaluated as substrates in stem cell culture. Upon screening diverse polymers, poly(methyl vinyl ether-alt-maleic anhydride) (PMVE-alt-MA) was able to support the long-term maintenance of two hESC lines, HUES1 and HUES9 (Brafman et al., 2010). A HTPS of 141 monomers and 909 unique polymers identified poly (HPhMA-co-HEMA), which stimulates the self-renewal of hESCs and hiPSCs (Celiz et al., 2015). Another HTPS on ~700 polymers identified a co-polymer of isobornyl methacrylate and tert-butylamino-ethyl methacrylate that supported myofibril organization in hESC-derived cardiomyocytes (Patel et al., 2015). Finally, small molecule chemical functional groups were screened in a 2D poly(ethyleneglycol) array to determine hits capable of inducing hMSC differentiation (Benoit et al., 2008) which were subsequently used to design 3D encapsulation materials. More complex designs have additionally been incorporated in HTPSs. For instance, Tewary et al reported the use of a micropatterned HTPS system in order to explore peri-gastrulation fate patterning in geometrically confined hPSC colonies treated with bone morphogenetic protein 4 (BMP4) (Tewary et al., 2019). Recently, the cultivation of 3D organoids and miniaturization of screening open up more opportunities for the understanding of the intricate relationship underpinning stem cells and their extracellular environment.
Disease modeling and drug discovery
hPSC-derived cells and organoids create physiologically and pathologically relevant platforms for drug discovery (Figure 3) (Friese et al., 2019). Zhou et al screened around 2000 compounds from the ToxCast library and discovered that a commonly used pesticide, propargite, induces pancreatic β-cell and dopamine neuron-specific death (Zhou et al., 2018). Amin et al combined CRISPR-based gene editing and HTPS to identify galunisertib, a TGF-β inhibitor, which rescued the loss of GLIS3, reversing pancreatic beta cell death (Amin et al., 2018).
Figure 3. Therapeutic applications of HTPS-based discoveries.
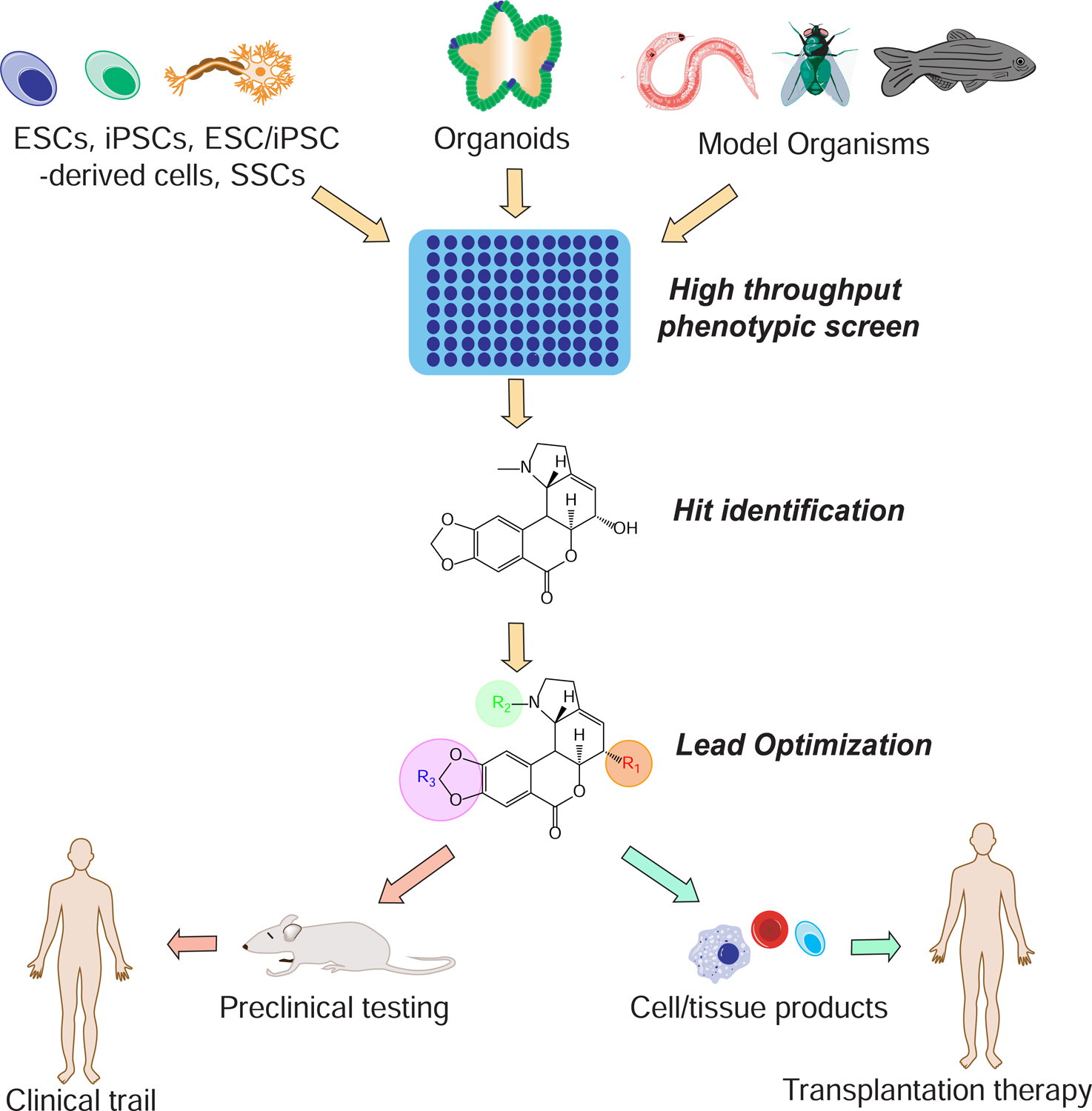
HTPSs can be adapted to include different biological systems including cells, organoid systems, and whole animals. Following the discovery of hit compounds, targets are validated and lead optimization is performed. Optimized compounds can eventually be used for cell-based therapy or be developed into a clinical drug, serving multiple therapeutic purposes.
More recently, stem cell-based HTPSs were applied for drug screening of infectious diseases. A hNPC based HTPS for anti-ZIKV drugs led to the identification of emricasan as a pan-caspase inhibitor that protects hNPCs, in addition to cyclin-dependent kinases and niclosamide that inhibit ZIKV replication (Xu et al., 2016). Zhou et al performed another HTPS and identified an anti-ZIKV compound, hippeastrine hydrobromide, that suppressed viral propagation when administered to adult mice with active ZIKV infection (Zhou et al., 2017). Recently, Yang et al reported a panel of hPSC-derived cells and organoids to study their permissiveness to SARS-CoV-2 infection (Yang et al., 2020). The hPSC-derived lung organoids were adapted to HTPS to screen 1280 FDA approved drugs. Three drug candidates, including imatinib, mycophenolic acid, and quinacrine dihydrochloride, were discovered to block SARS-CoV-2 entry both in vitro and in vivo (Han et al., 2020). In summary, these stem cell based HTPSs use cells closely resembling primary disease relevant cells/organs, which provides a new-generation platform for drug screening.
Whole animal screening
HTPS in a whole animal context has been gaining momentum with the development of robotic and automated imaging techniques. Preliminary hits identified using cell-based systems often do not exhibit a physiological effect upon in vivo administration, due to unfavorable drug absorption, distribution, metabolism, excretion, and toxicity (ADMET) properties. Caenorhabditis elegans (C. elegans), Drosophila melanogaster (D. melanogaster), and Danio rerio (D. rerio) are excellent animal models as they are inexpensive, easy to culture, and genetically tractable (Giacomotto and Segalat, 2010). D. rerio or zebrafish models have been especially relevant in the context of stem cell biology. For example, the Tg(isl1:GFP) transgenic zebrafish was utilized to discover the compound, D1, which inhibits the phosphorylation of ERK2 to promote stem cell renewal (Yin et al., 2017). Compounds including pentamidine, BML-267, and Vitamin D analogs have been reported to promote bone formation in zebrafish (Chen et al., 2017). With further advancement in detection technologies, whole organism screening could become highly valuable for disease modeling and drug discovery.
Target deconvolution
The major challenge associated with phenotypic screening approaches lies with target identification. The emergence of new technologies has made target deconvolution much more feasible than before.
Affinity-based approaches have been broadly used for target deconvolution (Figure 4). In brief, structure-activity relationship (SAR) analysis is employed to identify functional groups for linker attachment of the lead compound to functionalized beads, which allow for enrichment of the target(s) of the lead compound. For example, streptavidin-biotin based affinity purification identified nucleoside diphosphate kinase B as the target protein of stauprimide, a molecule that primes ESCs for differentiation (Zhu et al., 2009). Photoaffinity labeling (PAL) is a similar strategy which results in cross-linking of the target with the lead compound upon UV irradiation. A PAL-based approach was employed to discover tubulin as the molecular target of a series of compounds, 2,6-disubstituted 3H-imidazo[4,5-b]pyridines, which can be applied to treat dysferlinopathies in iPSC-derived myocytes (Takada et al., 2019). Furthermore, cellular thermal shift assay (CETSA) is an emerging approach established on the basis that many hit compounds increase target stability. Heat exposure degrades all the other proteins while the target remains intact and can be isolated and validated. For example, Marian et al reported the discovery of a macrolactam, BRDK25923209, from a HTPS as a potent modulator of the BAF (mammalian SW1/SNF) chromatin remodeling complex in mESCs and hence, a potential therapeutic option for HIV-1 latency reversal (Marian et al., 2018). Using CETSA, they demonstrated significant stabilization, and thus validation, of target proteins ARID1A, PBRM1, and LAMINB1 upon binding of BRDK25923209.
Figure 4. Biochemical strategies available for the target deconvolution.

Direct approaches typically include the isolation or characterization of the target protein bound to the compound of interest while indirect approaches involve the determination of changes in gene expression associated with the treatment of the identified compound.
In addition to direct deconvolution methods, transcriptional profiling, proteomics, phosphoproteomics, metabolic profiling, RNAi or CRISPR-Cas9 screens are also broadly applied to decipher genetic associations correlated with hit compounds. The genes discovered using these unbiased approaches are further subjected to knockout/knockdown or activated for further validation. For instance, Kimura et al reported the combined use of transcriptomics, namely RNA-sequencing and a siRNA screen, and phosphoproteomics to uncover the mechanism of action of AT7867 in regulating PP cell proliferation (Kimura et al., 2020). AT7867 was subsequently found to modulate the non-canonical WNT7B/PKC signaling pathway by inhibiting the phosphorylation of YY1, a multifunctional transcription factor regulating WNT7B expression.
The development of computer algorithms and machine learning applications has accelerated the possibility of sieving through the wealth of available biochemical information (Table 3). These approaches provide yet another strategy for target deconvolution while reducing the time and labor necessary to deconstruct the mechanism of action of a lead compound. Together, these biochemical, genetic, and computational target deconvolution approaches provide a powerful combination of tools to decode the biological targets and the mechanism of action of lead compounds.
Table 3.
Computational approaches for target identification
| Method | Feature |
|---|---|
| Protein-Ligand Interaction Fingerprints | The lead compound is docked to the binding pocket of a protein in energetically favorable binding poses and the free energy of binding of each pose is scored. |
| Chemical similarity ensemble approach | Proteins are related to each other based on the set-wise chemical similarity of their ligands. |
| Quantitative structure activity relationship (QSAR) | The biological activity of the lead compound is correlated with its physiological and chemical properties using algorithms or an artificial neural network using molecular descriptors including molecular similarity matrices derived from similarity calculations of parameters such as shape. |
| Proteochemometrics modeling | Proteochemometrics serves as an extension of QSAR and models the bioactivity of multiple ligands against multiple related targets, providing predictions on the bioactivity of compounds on untested targets. |
| Molecular dynamics/Binding affinity calculations | Molecular docking and dynamics (MD) simulations are used to elucidate ligand-target interactions and evaluate binding energies. |
Concluding remarks
The evolution of stem cell biology has been particularly fueled by HTPS-based discovery, which has revealed synthetic small molecules that influence key cell fate decisions. As a result, it has become increasingly facile to manipulate stem cells over the years, rendering them as capable systems for disease modeling, drug discovery, and cell-based therapeutics. However, stem cell-based systems are far from ideal. For instance, highly efficient differentiation protocols for most of cell types are still lacking. Moreover, in vitro cellular states do not accurately recapitulate their adult in vivo counterparts and often lack maturity. Over the years, improvements in cell-culturing techniques and a greater understanding of the stem cell niche via screens or otherwise, have helped to overcome this, at least in part. Additionally, efforts to mimic the cell state in the physiological environment have led to the invention of organoid and microfluidic systems which reveal the intricacies of an integrated system. Overall, HTPSs will develop novel chemical tools to generate mature functional cells/organoids that can be used for replacement therapy, disease modeling and drug discovery.
Significance.
Modulating key cell fate decisions and elucidating complex biological pathways remain some of the major challenges in the field of stem cell biology. Here, we elaborate on the value of HTPS as a tool to uncover novel chemical candidates that regulate stem cell fate and/or mimic the extracellular stem cell niche and to deconvolute intricate mechanisms. Recent developments in HTPS technologies pave the way for significant breakthroughs in stem cell biology pertaining to drug discovery, disease modeling, and regenerative medicine.
Highlights.
HTPS revealed modulators of self-renewal, reprogramming, and differentiation.
Modulators mimicking the stem cell niche have been uncovered using HTPS.
Cellular or animal systems can be adapted for HTPS in drug discovery.
Target validation strategies elucidating complex biological pathways are available.
Acknowledgements
This work was supported by NIDDK (R01 DK116075-01A1, R01 DK119667-01A1, R01 DK124463, 1 DP3DK111907-01, and U01 DK127777-01), and American Heart Association (18CSA34080171)..
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare no competing interests.
References
- Ali D, Hamam R, Alfayez M, Kassem M, Aldahmash A, and Alajez NM (2016). Epigenetic Library Screen Identifies Abexinostat as Novel Regulator of Adipocytic and Osteoblastic Differentiation of Human Skeletal (Mesenchymal) Stem Cells. Stem Cell Transl Med 5, 1036–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin S, Cook B, Zhou T, Ghazizadeh Z, Lis R, Zhang T, Khalaj M, Crespo M, Perera M, Xiang JZ, et al. (2018). Discovery of a drug candidate for GLIS3-associated diabetes. Nat Commun 9, 2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araoka T, Mae S, Kurose Y, Uesugi M, Ohta A, Yamanaka S, and Osafune K (2014). Efficient and Rapid Induction of Human iPSCs/ESCs into Nephrogenic Intermediate Mesoderm Using Small Molecule-Based Differentiation Methods. Plos One 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arulmozhivarman G, Krater M, Wobus M, Friedrichs J, Bejestani EP, Muller K, Lambert K, Alexopoulou D, Dahl A, Stoter M, et al. (2017). Zebrafish In-Vivo Screening for Compounds Amplifying Hematopoietic Stem and Progenitor Cells: - Preclinical Validation in Human CD34+Stem and Progenitor Cells. Sci Rep-Uk 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astuti Y, Kramer AC, Blake AL, Blazar BR, Tolar J, Taisto ME, and Lund TC (2017). A Functional Bioluminescent Zebrafish Screen for Enhancing Hematopoietic Cell Homing. Stem Cell Rep 8, 177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaric I, Gokhale PJ, Jones M, Glen A, Baker D, and Andrews PW (2010). Novel regulators of stem cell fates identified by a multivariate phenotype screen of small compounds on human embryonic stem cell colonies. Stem Cell Research 5, 104–119. [DOI] [PubMed] [Google Scholar]
- Benoit DSW, Schwartz MP, Durney AR, and Anseth KS (2008). Small functional groups for controlled differentiation of hydrogel-encapsulated human mesenchymal stem cells. Nat Mater 7, 816–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boitano AE (2011). Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells (September, pg 1345, 2010). Science 332, 664–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiak M, Maehr R, Chen S, Chen AE, Tang W, Fox JL, Schreiber SL, and Melton DA (2009). Small molecules efficiently direct endodermal differentiation of mouse and human embryonic stem cells. Cell Stem Cell 4, 348–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brafman DA, Chang CW, Fernandez A, Willert K, Varghese S, and Chien S (2010). Long-term human pluripotent stem cell self-renewal on synthetic polymer surfaces. Biomaterials 31, 9135–9144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brafman DA, Phung C, Kumar N, and Willert K (2013). Regulation of endodermal differentiation of human embryonic stem cells through integrin-ECM interactions. Cell Death Differ 20, 369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celiz AD, Smith JGW, Patel AK, Hook AL, Rajamohan D, George VT, Flatt L, Patel MJ, Epa VC, Singh T, et al. (2015). Discovery of a Novel Polymer for Human Pluripotent Stem Cell Expansion and Multilineage Differentiation. Adv Mater 27, 4006–4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Qi YC, Mica Y, Lee G, Zhang XJ, Niu L, Bilsland J, Cao LS, Stevens E, Whiting P, et al. (2012). Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nat Biotechnol 30, 715–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbord J, Poydenot P, Bonnefond C, Feyeux M, Casagrande F, Brinon B, Francelle L, Auregan G, Guillermier M, Cailleret M, et al. (2013). High Throughput Screening for Inhibitors of REST in Neural Derivatives of Human Embryonic Stem Cells Reveals a Chemical Compound that Promotes Expression of Neuronal Genes. Stem Cells 31, 1816–1828. [DOI] [PubMed] [Google Scholar]
- Chen JR, Lai YH, Tsai JJ, and Hsiao CD (2017). Live Fluorescent Staining Platform for Drug-Screening and Mechanism-Analysis in Zebrafish for Bone Mineralization. Molecules 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Borowiak M, Fox JL, Maehr R, Osafune K, Davidow L, Lam K, Peng LF, Schreiber SL, Rubin LL, et al. (2009). A small molecule that directs differentiation of human ESCs into the pancreatic lineage. Nature Chemical Biology 5, 258–265. [DOI] [PubMed] [Google Scholar]
- Chen S, Do JT, Zhang Q, Yao S, Yan F, Peters EC, Scholer HR, Schultz PG, and Ding S (2006). Self-renewal of embryonic stem cells by a small molecule. Proc Natl Acad Sci U S A 103, 17266–17271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YS, Wu BB, Lin JX, Yu DS, Du XT, Sheng ZX, Yu YK, An CR, Zhang XA, Li QK, et al. (2020). High-Resolution Dissection of Chemical Reprogramming from Mouse Embryonic Fibroblasts into Fibrocartilaginous Cells. Stem Cell Rep 14, 478–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook B, Rafiq R, Lee H, Banks KM, El-Debs M, Chiaravalli J, Glickman JF, Das BC, Chen SB, and Evans T (2019). Discovery of a Small Molecule Promoting Mouse and Human Osteoblast Differentiation via Activation of p38 MAPK-beta. Cell Chem Biol 26, 926–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham TJ, and Duester G (2015). Mechanisms of retinoic acid signalling and its roles in organ and limb development. Nat Rev Mol Cell Biol 16, 110–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damoiseaux R, Sherman SP, Alva JA, Peterson C, and Pyle AD (2009). Integrated chemical genomics reveals modifiers of survival in human embryonic stem cells. Stem Cells 27, 533–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbordes SC, Placantonakis DG, Ciro A, Socci ND, Lee G, Djaballah H, and Studer L (2008). High-throughput screening assay for the identification of compounds regulating self-renewal and differentiation in human embryonic stem cells. Cell Stem Cell 2, 602–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi A, Sevinc K, Gurhan Sevinc G, Cribbs AP, Philpott M, Uyulur F, Morova T, Dunford JE, Goklemez S, Ari S, et al. (2019). Bromodomain inhibition of the coactivators CBP/EP300 facilitate cellular reprogramming. Nat Chem Biol 15, 519–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, and Kaufman MH (1981). Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156. [DOI] [PubMed] [Google Scholar]
- Fares I, Chagraoui J, Gareau Y, Gingras S, Ruel R, Mayotte N, Csaszar E, Knapp DJHF, Miller P, Ngom M, et al. (2014). Pyrimidoindole derivatives are agonists of human hematopoietic stem cell self-renewal. Science 345, 1509–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friese A, Ursu A, Hochheimer A, Scholer HR, Waldmann H, and Bruder JM (2019). The Convergence of Stem Cell Technologies and Phenotypic Drug Discovery. Cell Chem Biol 26, 1050–1066. [DOI] [PubMed] [Google Scholar]
- Ghazizadeh Z, Kao DI, Amin S, Cook B, Rao S, Zhou T, Zhang T, Xiang ZY, Kenyon R, Kaymakcalan O, et al. (2017). ROCKII inhibition promotes the maturation of human pancreatic beta-like cells. Nat Commun 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomotto J, and Segalat L (2010). High-throughput screening and small animal models, where are we? Brit J Pharmacol 160, 204–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong SQ, Han XW, Li XH, Yang J, He XB, and Si SY (2016). Development of a High-Throughput Screening Strategy for Upregulators of the OPG/RANKL Ratio with the Potential for Antiosteoporosis Effects. J Biomol Screen 21, 738–748. [DOI] [PubMed] [Google Scholar]
- Gonzalez R, Jennings LL, Knuth M, Orth AP, Klock HE, Ou W, Feuerhelm J, Hull MV, Koesema E, Wang YP, et al. (2010). Screening the mammalian extracellular proteome for regulators of embryonic human stem cell pluripotency. P Natl Acad Sci USA 107, 3552–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagedorn EJ, Durand EM, Fast EM, and Zon LI (2014). Getting more for your marrow: Boosting hematopoietic stem cell numbers with PGE(2). Exp Cell Res 329, 220–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han XW, Gong SQ, Li N, Wang X, Liu P, Xu YN, He XB, Jiang W, and Si SY (2019). A Novel Small Molecule Which Increases Osteoprotegerin Expression and Protects Against Ovariectomy-Related Bone Loss in Rats. Front Pharmacol 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Duan X, Yang L, Nilsson-Payant BE, Wang P, Duan F, Tang X, Yaron TM, Zhang T, Uhl S, et al. (2020). Identification of SARS-CoV-2 Inhibitors using Lung and Colonic Organoids. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Headley KM, Kedziora KM, Alejo A, Lai EZ, Purvis JE, and Hathaway NA (2019). Chemical screen for epigenetic barriers to single allele activation of Oct4. Stem Cell Res 38, 101470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann T, and Gastreich M (2019). The next level in chemical space navigation: going far beyond enumerable compound libraries. Drug Discov Today 24, 1148–1156. [DOI] [PubMed] [Google Scholar]
- Hou PP, Li YQ, Zhang X, Liu C, Guan JY, Li HG, Zhao T, Ye JQ, Yang WF, Liu K, et al. (2013). Pluripotent Stem Cells Induced from Mouse Somatic Cells by Small-Molecule Compounds. Science 341, 651–654. [DOI] [PubMed] [Google Scholar]
- Huang AH, Motlekar NA, Stein A, Diamond SL, Shore EM, and Mauck RL (2008). High-throughput screening for modulators of mesenchymal stem cell chondrogenesis. Ann Biomed Eng 36, 1909–1921. [DOI] [PubMed] [Google Scholar]
- Huangfu DW, Maehr R, Guo WJ, Eijkelenboom A, Snitow M, Chen AE, and Melton DA (2008a). Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol 26, 795–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu DW, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, and Melton DA (2008b). Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol 26, 1269–1275. [DOI] [PubMed] [Google Scholar]
- Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, Egli D, Loh KM, Carter AC, Di Giorgio FP, Koszka K, et al. (2009). A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell 5, 491–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Hara H, Takeda N, Naito AT, Nomura S, Kondo M, Hata Y, Uchiyama M, Morita H, and Komuro I (2019). Characterization of a small molecule that promotes cell cycle activation of for human induced pluripotent stem cell-derived cardiomyocytes. J Mol Cell Cardiol 128, 90–95. [DOI] [PubMed] [Google Scholar]
- Jung P, Sato T, Merlos-Suarez A, Barriga FM, Iglesias M, Rossell D, Auer H, Gallardo M, Blasco MA, Sancho E, et al. (2011). Isolation and in vitro expansion of human colonic stem cells. Nat Med 17, 1225–1227. [DOI] [PubMed] [Google Scholar]
- Kabeya T, Qiu SM, Hibino M, Nagasaki M, Kodama N, Iwao T, and Matsunaga T (2018). Cyclic AMP Signaling Promotes the Differentiation of Human Induced Pluripotent Stem Cells into Intestinal Epithelial Cells. Drug Metab Dispos 46, 1411–1419. [DOI] [PubMed] [Google Scholar]
- Kimura A, Toyoda T, Iwasaki M, Hirama R, and Osafune K (2020). Combined Omics Approaches Reveal the Roles of Non-canonical WNT7B Signaling and YY1 in the Proliferation of Human Pancreatic Progenitor Cells. Cell Chem Biol. [DOI] [PubMed] [Google Scholar]
- Kimura A, Toyoda T, Nishi Y, Nasu M, Ohta A, and Osafune K (2017). Small molecule AT7867 proliferates PDX1-expressing pancreatic progenitor cells derived from human pluripotent stem cells. Stem Cell Research 24, 61–68. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Nishimura H, Kudo N, Osada H, and Yoshida M (2020). A novel GSK3 inhibitor that promotes self-renewal in mouse embryonic stem cells. Biosci Biotech Bioch. [DOI] [PubMed] [Google Scholar]
- Koepsel JT, Brown PT, Loveland SG, Li WJ, and Murphy WL (2012). Combinatorial screening of chemically defined human mesenchymal stem cell culture substrates. J Mater Chem 22, 19474–19481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Toyoda T, Ito R, Funato M, Hosokawa Y, Matsui S, Sudo T, Nakamura M, Okada C, Zhuang XT, et al. (2017). Identification of a small molecule that facilitates the differentiation of human iPSCs/ESCs and mouse embryonic pancreatic explants into pancreatic endocrine cells. Diabetologia 60, 1454–1466. [DOI] [PubMed] [Google Scholar]
- Korostylev A, Mahaddalkar PU, Keminer O, Hadian K, Schorpp K, Gribbon P, and Lickert H (2017). A high-content small molecule screen identifies novel inducers of definitive endoderm. Mol Metab 6, 640–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Lee J, and Cho YS (2018). Peroxisome Proliferator-Activated Receptor alpha Agonist and Its Target Nanog Cooperate to Induce Pluripotency. J Clin Med 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, An J, Kang TM, De D, and Kim KK (2019). Discovery of Natural Compounds Promoting Cardiomyocyte Differentiation. Stem Cells Dev 28, 13–27. [DOI] [PubMed] [Google Scholar]
- Li PL, Lahvic JL, Binder V, Pugach EK, Riley EB, Tamplin OJ, Panigrahy D, Bowman TV, Barrett FG, Heffner GC, et al. (2015a). Epoxyeicosatrienoic acids enhance embryonic haematopoiesis and adult marrow engraftment. Nature 523, 468–U203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zuo XH, Jing JZ, Ma YT, Wang JM, Liu DF, Zhu JL, Du XM, Xiong L, Du YY, et al. (2015b). Small-Molecule-Driven Direct Reprogramming of Mouse Fibroblasts into Functional Neurons. Cell Stem Cell 17, 195–203. [DOI] [PubMed] [Google Scholar]
- Long Y, Wang M, Gu H, and Xie X (2015). Bromodeoxyuridine promotes full-chemical induction of mouse pluripotent stem cells. Cell Res 25, 1171–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumelsky N (2014). Small Molecules Convert Fibroblasts into Islet-like Cells Avoiding Pluripotent State. Cell Metab 19, 551–552. [DOI] [PubMed] [Google Scholar]
- Lyssiotis CA, Foreman RK, Staerk J, Garcia M, Mathur D, Markoulaki S, Hanna J, Lairson LL, Charette BD, Bouchez LC, et al. (2009). Reprogramming of murine fibroblasts to induced pluripotent stem cells with chemical complementation of Klf4. Proc Natl Acad Sci U S A 106, 8912–8917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma TH, Li J, Xu Y, Yu C, Xu T, Wang HX, Liu K, Cao N, Nie BM, Zhu SY, et al. (2015). Atg5-independent autophagy regulates mitochondrial clearance and is essential for iPSC reprogramming. Nat Cell Biol 17, 1379–1387. [DOI] [PubMed] [Google Scholar]
- Marian CA, Stoszko M, Wang LL, Leighty MW, de Crignis E, Maschinot CA, Gatchalian J, Carter BC, Chowdhury B, Hargreaves DC, et al. (2018). Small Molecule Targeting of Specific BAF (mSWI/SNF) Complexes for HIV Latency Reversal. Cell Chem Biol 25, 1443–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR (1981). Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A 78, 7634–7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei YQ, Pan ZF, Chen WT, Xu MH, Zhu DY, Yu YP, and Lou YJ (2016). A Flavonoid Compound Promotes Neuronal Differentiation of Embryonic Stem Cells via PPAR-beta Modulating Mitochondrial Energy Metabolism. PLoS One 11, e0157747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami I, Yamada K, Otsuji TG, Yamamoto T, Shen Y, Otsuka S, Kadota S, Morone N, Barve M, Asai Y, et al. (2012). A Small Molecule that Promotes Cardiac Differentiation of Human Pluripotent Stem Cells under Defined, Cytokine- and Xeno-free Conditions. Cell Rep 2, 1448–1460. [DOI] [PubMed] [Google Scholar]
- Miyabayashi T, Teo JL, Yamamoto M, McMillan M, Nguyen C, and Kahn M (2007). Wnt/beta-catenin/CBP signaling maintains long-term murine embryonic stem cell pluripotency. Proc Natl Acad Sci U S A 104, 5668–5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed TMA, Stone NR, Berry EC, Radzinsky E, Huang Y, Pratt K, Ang YS, Yu PZ, Wang HX, Tang SB, et al. (2017). Chemical Enhancement of In Vitro and In Vivo Direct Cardiac Reprogramming. Circulation 135, 978–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North TE, Goessling W, Peeters M, Li PL, Ceol C, Lord AM, Weber GJ, Harris J, Cutting CC, Huang P, et al. (2009). Hematopoietic Stem Cell Development Is Dependent on Blood Flow. Cell 137, 736–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North TE, Goessling W, Walkley CR, Lengerke C, Kopani KR, Lord AM, Weber GJ, Bowman TV, Jang IH, Grosser T, et al. (2007). Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature 447, 1007–U1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AK, Celiz AD, Rajamohan D, Anderson DG, Langer R, Davies MC, Alexander MR, and Denning C (2015). A defined synthetic substrate for serum-free culture of human stem cell derived cardiomyocytes with improved functional maturity identified using combinatorial materials microarrays. Biomaterials 61, 257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfisterer U, Ek F, Lang S, Soneji S, Olsson R, and Parmar M (2016). Small molecules increase direct neural conversion of human fibroblasts. Sci Rep-Uk 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, and Marshak DR (1999). Multilineage potential of adult human mesenchymal stem cells. Science 284, 143–147. [DOI] [PubMed] [Google Scholar]
- Rezania A, Bruin JE, Arora P, Rubin A, Batushansky I, Asadi A, O’Dwyer S, Quiskamp N, Mojibian M, Albrecht T, et al. (2014). Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat Biotechnol 32, 1121–1133. [DOI] [PubMed] [Google Scholar]
- Sadek H, Hannack B, Choe E, Wang J, Latif S, Garry MG, Garry DJ, Longgood J, Frantz DE, Olson EN, et al. (2008). Cardiogenic small molecules that enhance myocardial repair by stem cells. P Natl Acad Sci USA 105, 6063–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano D, Shiraki N, Kikawa K, Yamazoe T, Kataoka M, Umeda K, Araki K, Mao D, Matsumoto S, Nakagata N, et al. (2014). VMAT2 identified as a regulator of late-stage beta-cell differentiation. Nature Chemical Biology 10, 141–148. [DOI] [PubMed] [Google Scholar]
- Selvaraj S, Mondragon-Gonzalez R, Xu B, Magli A, Kim H, Laine J, Kiley J, Mckee H, Rinaldi F, Aho J, et al. (2019). Screening identifies small molecules that enhance the maturation of human pluripotent stem cell-derived myotubes. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan J, Schwartz RE, Ross NT, Logan DJ, Thomas D, Duncan SA, North TE, Goessling W, Carpenter AE, and Bhatia SN (2013). Identification of small molecules for human hepatocyte expansion and iPS differentiation. Nature Chemical Biology 9, 514–U577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Desponts C, Do JT, Hahm HS, Scholer HR, and Ding S (2008). Induction of Pluripotent Stem Cells from Mouse Embryonic Fibroblasts by Oct4 and Klf4 with Small-Molecule Compounds. Cell Stem Cell 3, 568–574. [DOI] [PubMed] [Google Scholar]
- Staerk J, Lyssiotis CA, Medeiro LA, Bollong M, Foreman RK, Zhu ST, Garcia M, Gao Q, Bouchez LC, Lairson LL, et al. (2011). Pan-Src Family Kinase Inhibitors Replace Sox2 during the Direct Reprogramming of Somatic Cells. Angew Chem Int Edit 50, 5733–5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Dong Z, Jin T, Ang KH, Huang M, Haston KM, Peng J, Zhong TP, Finkbeiner S, Weiss WA, et al. (2013). Imaging-based chemical screening reveals activity-dependent neural differentiation of pluripotent stem cells. Elife 2, e00508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada H, Kaieda A, Tawada M, Nagino T, Sasa K, Oikawa T, Oki A, Sameshima T, Miyamoto K, Miyamoto M, et al. (2019). Identification of 2,6-Disubstituted 3H-Imidazo[4,5-b]pyridines as Therapeutic Agents for Dysferlinopathies through Phenotypic Screening on Patient-Derived Induced Pluripotent Stem Cells. J Med Chem 62, 9175–9187. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, and Yamanaka S (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872. [DOI] [PubMed] [Google Scholar]
- Takahashi K, and Yamanaka S (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Lord B, Schulze PC, Fryer RM, Sarang SS, Gullans SR, and Lee RT (2003). Ascorbic acid enhances differentiation of embryonic stem cells into cardiac myocytes. Circulation 107, 1912–1916. [DOI] [PubMed] [Google Scholar]
- Tewary M, Dziedzicka D, Ostblom J, Prochazka L, Shakiba N, Heydari T, Aguilar-Hidalgo D, Woodford C, Piccinini E, Becerra-Alonso D, et al. (2019). High-throughput micropatterning platform reveals Nodal-dependent bisection of peri-gastrulation-associated versus preneurulation-associated fate patterning. Plos Biol 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, and Jones JM (1998). Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147. [DOI] [PubMed] [Google Scholar]
- Tsai SY, Maass K, Lu J, Fishman GI, Chen S, and Evans T (2015). Efficient Generation of Cardiac Purkinje Cells from ESCs by Activating cAMP Signaling. Stem Cell Reports 4, 1089–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursu A, Scholer HR, and Waldmann H (2017). Small-molecule phenotypic screening with stem cells. Nat Chem Biol 13, 560–563. [DOI] [PubMed] [Google Scholar]
- Vendrell M, Park SJ, Chandran Y, Lee CL, Ha HH, Kang NY, Yun SW, and Chang YT (2012). A fluorescent screening platform for the rapid evaluation of chemicals in cellular reprogramming. Stem Cell Res 9, 185–191. [DOI] [PubMed] [Google Scholar]
- Wang DY, Hu YZ, Kong SS, Yu YP, Zhu DY, and Lou YJ (2011). Promoting effects of isobavachin on neurogenesis of mouse embryonic stem cells were associated with protein prenylation. Acta Pharmacol Sin 32, 425–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wang H, Wu J, Zhu D, Zhang X, Ou L, Yu Y, and Lou Y (2009). Enhanced co-expression of beta-tubulin III and choline acetyltransferase in neurons from mouse embryonic stem cells promoted by icaritin in an estrogen receptor-independent manner. Chem Biol Interact 179, 375–385. [DOI] [PubMed] [Google Scholar]
- Willems E, Spiering S, Davidovics H, Lanier M, Xia ZB, Dawson M, Cashman J, and Mercola M (2011). Small-Molecule Inhibitors of the Wnt Pathway Potently Promote Cardiomyocytes From Human Embryonic Stem Cell-Derived Mesoderm. Circ Res 109, 360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Ding S, Ding G, Gray NS, and Schultz PG (2004). Small molecules that induce cardiomyogenesis in embryonic stem cells. J Am Chem Soc 126, 1590–1591. [DOI] [PubMed] [Google Scholar]
- Xu M, Lee EM, Wen Z, Cheng Y, Huang WK, Qian X, Tcw J, Kouznetsova J, Ogden SC, Hammack C, et al. (2016). Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat Med 22, 1101–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang LL, Han YL, Nilsson-Payant BE, Gupta V, Wang PF, Duan XH, Tang XM, Zhu JJ, Zhao ZP, Jaffre F, et al. (2020). A Human Pluripotent Stem Cell-based Platform to Study SARS-CoV-2 Tropism and Model Virus Infection in Human Cells and Organoids. Cell Stem Cell 27, 125–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Liu B, Xu J, Wang JL, Wu J, Shi C, Xu YX, Dong JB, Wang CY, Lai WF, et al. (2017). Derivation of Pluripotent Stem Cells with In Vivo Embryonic and Extraembryonic Potency. Cell 169, 243–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Ge J, Zhang X, Cheng L, Zhang Z, He S, Wang Y, Lin H, Yang W, Liu J, et al. (2016). Pluripotent stem cells induced from mouse neural stem cells and small intestinal epithelial cells by small molecule compounds. Cell Res 26, 34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C, Fufa T, Chandrasekar G, Aeluri M, Zaky V, Abdelhady S, Rodriguez AB, Jakobsson J, Varnoosfaderani FS, Mahalingam J, et al. (2017). Phenotypic Screen Identifies a Small Molecule Modulating ERK2 and Promoting Stem Cell Proliferation. Front Pharmacol 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JY, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920. [DOI] [PubMed] [Google Scholar]
- Yuan X, Wan H, Zhao X, Zhu S, Zhou Q, and Ding S (2011). Brief report: combined chemical treatment enables Oct4-induced reprogramming from mouse embryonic fibroblasts. Stem Cells 29, 549–553. [DOI] [PubMed] [Google Scholar]
- Zhang L, Yin JC, Yeh H, Ma NX, Lee G, Chen XA, Wang YM, Lin L, Chen L, Jin P, et al. (2015). Small Molecules Efficiently Reprogram Human Astroglial Cells into Functional Neurons. Cell Stem Cell 17, 735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JX, Su P, Li D, Tsang S, Duan EK, and Wang F (2010). High-Efficiency Induction of Neural Conversion in Human ESCs and Human Induced Pluripotent Stem Cells with a Single Chemical Inhibitor of Transforming Growth Factor Beta Superfamily Receptors. Stem Cells 28, 1741–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Kim TW, Chong CN, Tan L, Amin S, Sadat Badieyan Z, Mukherjee S, Ghazizadeh Z, Zeng H, Guo M, et al. (2018). A hPSC-based platform to discover gene-environment interactions that impact human beta-cell and dopamine neuron survival. Nat Commun 9, 4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Tan L, Cederquist GY, Fan Y, Hartley BJ, Mukherjee S, Tomishima M, Brennand KJ, Zhang Q, Schwartz RE, et al. (2017). High-Content Screening in hPSC-Neural Progenitors Identifies Drug Candidates that Inhibit Zika Virus Infection in Fetal-like Organoids and Adult Brain. Cell Stem Cell 21, 274–283 e275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Ambasudhan R, Sun W, Kim HJ, Talantova M, Wang XJ, Zhang ML, Zhang Y, Laurent T, Parker J, et al. (2014). Small molecules enable OCT4-mediated direct reprogramming into expandable human neural stem cells. Cell Research 24, 126–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Li W, Zhou H, Wei W, Ambasudhan R, Lin T, Kim J, Zhang K, and Ding S (2010). Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell 7, 651–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu ST, Wurdak H, Wang J, Lyssiotis CA, Peters EC, Cho CY, Wu X, and Schultz PG (2009). A Small Molecule Primes Embryonic Stem Cells for Differentiation. Cell Stem Cell 4, 416–426. [DOI] [PubMed] [Google Scholar]


