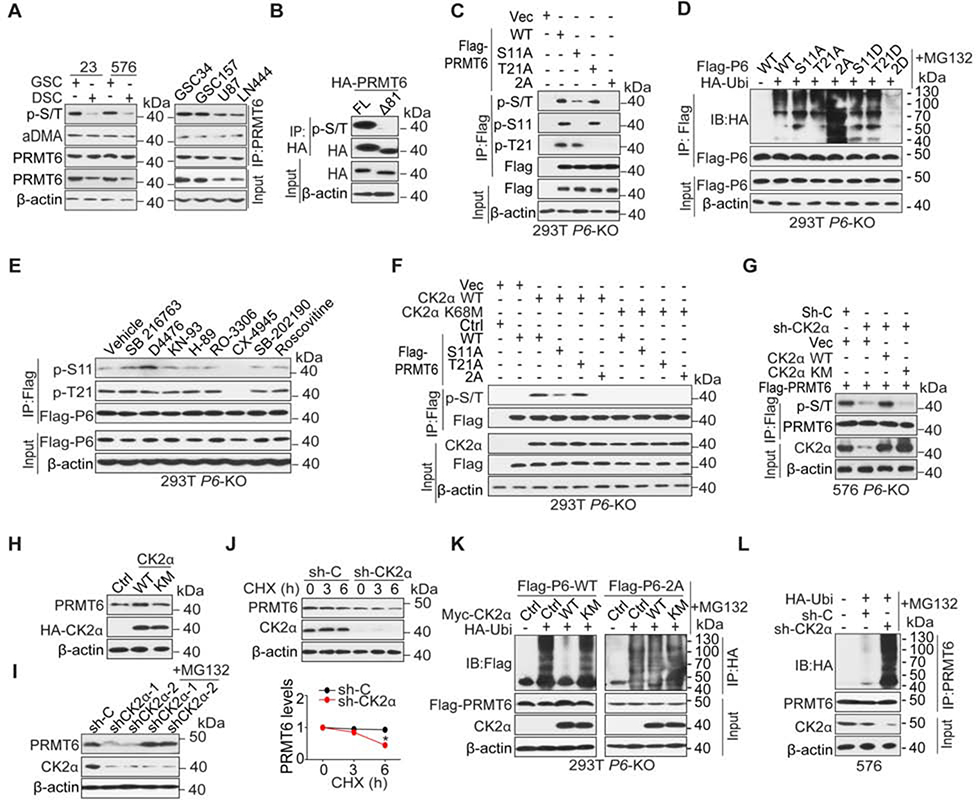
Figure 5. CK2α Stabilizes PRMT6 Protein through Phosphorylation of PRMT6.
(A) IP-IB and IB using indicated antibodies in GSCs, their corresponding differentiated glioma cells (DSCs), and glioma cell lines.
(B) IP-IB and IB using indicated antibodies in GSC576 cells expressing a full-length (FL) or N-terminal deletion mutant (Δ81) of HA-PRMT6.
(C-F) IP-IB and IB for indicated proteins in 293T/PRMT6 KO cells with indicated modifications or treatments.
(G) IP-IB and IB for p-S/T of PRMT6 in GSC576/PRMT6 KO cells that were transduced with indicated plasmids. sh-CK2α targets 3’UTR of CSNK2A1 mRNA, and GSC576 cells were collected after treatment with MG132 for 6 h.
(H) IB for PRMT6 in 293T cells that were transduced with indicated plasmids.
(I) IB for PRMT6 and CK2α in GSC576 cells with indicated modifications.
(J) IB for PRMT6 and CK2α in GSC23/CK2α KD or sh-C cells that were treated with 50 μg/ml cycloheximide (CHX) for the indicated times. Band intensities of PRMT6 proteins were quantified and the results were expressed as PRMT6 levels relative to untreated cells. Error bars, ± SEM, n=3. Two-tailed Student’s t-test. *, p < 0.05;
(K and L) IP-IB and IB for ubiquitination of PRMT6 in 293T P6-KO (K) and GSC576 (L) cells with indicated modifications and treatments.
Data are representative of two to three independent experiments with similar results.
See also Figure S5.