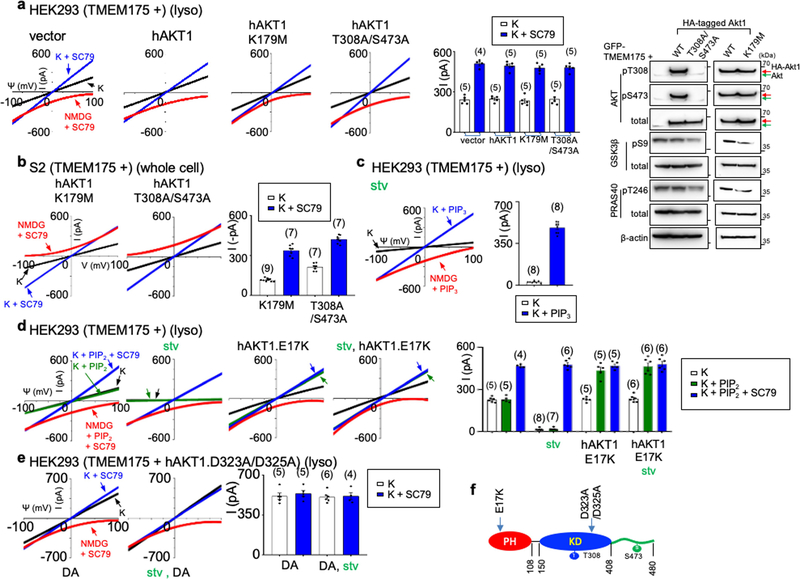
Extended Data Fig. 6 |. AKT determines the sensitivity of TMEM175 to starvation and activation by phosphatidylinositol lipids.
a–e, HEK293T cells (a, c–e) (lysosomal recording) or S2 cells (b) (whole-cell recording) were transfected with human TMEM175 with or without cotransfection of wild-type or mutant human AKT1. a, b, Experiments demonstrating that AKT with mutations in the kinase domain (K179M alone, T308A/S473A double mutant) are able to support ITMEM175 as well as wild-type AKT. Extended Data Figure 2 provides a comparison with those from S2 cells transfected with TMEM175 without human AKT1 cotransfection, in which very little currents were generated. A summary of current amplitudes (at −100 mV) is given in the bar graphs. The right panel in a shows the phosphorylation status of transfected AKT and AKT targets GSK3β and PRAS40. The human AKT1 used for transfection has an HA tag and a linker sequence, and can be distinguished from the endogenous AKT (indicated by green arrows) because of its slightly higher molecular weight. For gel source data, see Supplementary Fig. 1. In c–e, stv indicates cells starved in HBSS medium for 2 h before lysosomal dissection for recording. c, PIP3 (1 μM) was applied to the recording bath during some of the recordings (blue and red traces). d, e, ITMEM175 recorded from lysosomes with or without cotransfection of the E17K (d) or the D323A/D325A (DA) (e) AKT1 mutants, with or without starvation. PIP2 in d was applied at 1 μM. Data are mean ± s.e.m. Numbers of recordings are in parentheses. Arrows are used to indicate curves that overlap and are not easily distinguished. f, Positions of the AKT1 mutations used in this figure. We used AKT mutants to further test the nonessential role of kinase activity in ITMEM175. AKT1 with mutation at K179 (a key residue in the ATP- and Mg2+-binding pocket; K179M), or at T308 and S473 (residues key to AKT kinase activation; T308A/S473A) no longer phosphorylates its targets18,81. ITMEM175 from cells co-expressing the mutants, however, was similar to that co-expressing wild-type AKT (a). More importantly, when co-expressed with human TMEM175 in S2 cells, the kinase-inactive mutants fully supported ITMEM175 in a manner indistinguishable from that of the wild type (b). We then tested whether AKT conformational changes are sufficient to activate the channel. Under physiological conditions, PIP3 generated on the plasma membrane opens AKT by binding to the PH domain. In lysosomes from starved cells, PIP3 bath application activated ITMEM175 (c). The E17K mutation in the PH domain—commonly found in cancers—switches the lipid sensitivity of AKT from PIP3 to PIP2, a lipid that is abundant even during starvation82. Notably, ITMEM175 from cells cotransfected with AKT(E17K) was resistant to starvation, and, unlike that without cotransfection, was activated by PIP2 (d). Thus, AKT also determines the lipid sensitivity of TMEM175. The double mutation D323A/D325A of AKT1 disrupts the locking of the PH domain to the kinase domain and keeps AKT in a constitutively open state23,83. ITMEM175 from cells co-expressing this mutant was resistant to starvation, maximally activated and not further activated by SC79 (e).