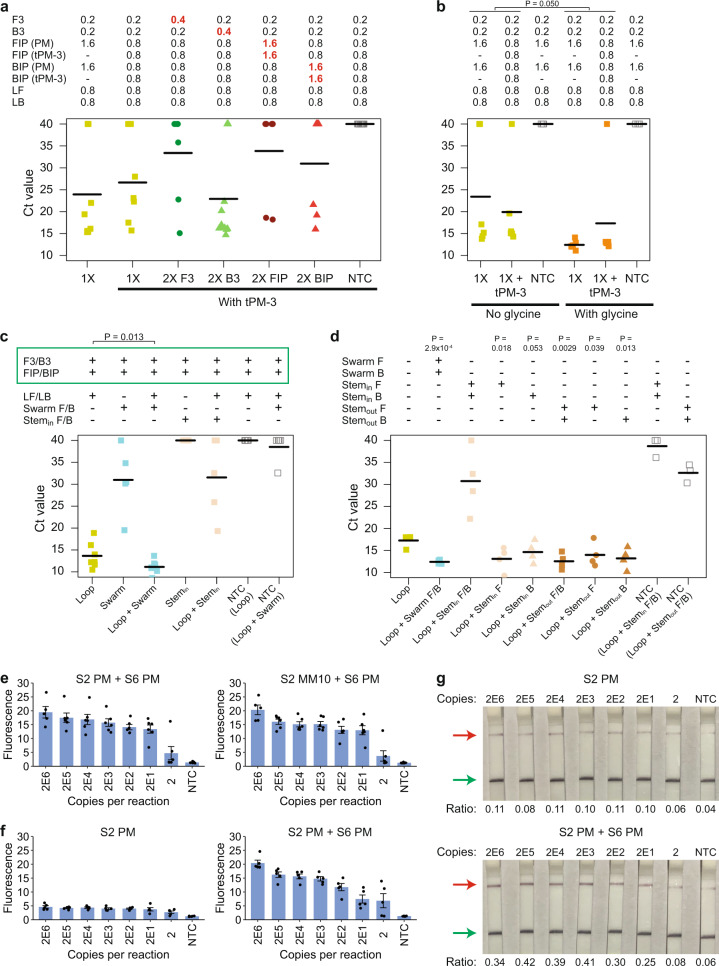
Fig. 4. Methods to improve sensitivity of LAMP.
a Strip chart showing how LAMP sensitivity was affected by the concentration of primers used. We tested different concentrations of displacement primers and internal primers. RT-LAMP was performed at 65 °C in a real-time instrument with 20 copies of RNA template corresponding to the S-gene of SARS-CoV-2. The black horizontal bars among the data points in the strip chart represent the mean (n = 7 [1X, 2X F3, 2X FIP, 2X BIP], 10 [1X without tPM], 12 [2X B3], or 15 [NTC] biological replicates). b Strip chart showing how LAMP sensitivity was altered by 0.1 M glycine. RT-LAMP was performed at 65 °C with 20 copies of RNA template. The black horizontal bars among the data points in the strip chart represent the mean (n = 3 [NTC] or 6 [with template] biological replicates). P-value was calculated using one-sided Student’s t-test. c Strip chart showing how LAMP sensitivity was altered by the use of swarm or stem primers. The green box demarcates the four core primers, which were included in every experiment. The concentrations of each displacement primer, internal primer, loop primer, swarm primer, and stem primer were 0.2, 1.6, 0.8, 1.6, and 1.6 µM, respectively. RT-LAMP was performed at 65 °C with 20,000 copies of RNA template. The black horizontal bars among the data points in the strip chart represent the mean (n = 5 [swarm, stemin, loop + stemin, NTC for loop + swarm] or 7 [loop, loop + swarm, NTC for loop] biological replicates). P-value was calculated using one-sided Student’s t-test. d Further dissection of stem primers. The strip chart shows the impact of various stem primers on LAMP sensitivity. Here, every reaction contained the displacement primers (0.2 µM each), internal primers (1.6 µM each), and loop primers (0.8 µM each). Furthermore, it could also contain either two additional swarm primers or one or two additional stem primers (with the concentration of each extra primer being 1.6 µM). RT-LAMP was performed at 65 °C with 20,000 copies of RNA template. The black horizontal bars among the data points in the strip chart represent the mean (n = 3 [NTC] or 4 [with template] biological replicates). P-values were calculated using one-sided Student’s t-test. e Analytical LoD for enAsCas12a complexed with both the S6 gRNA and either the PM or the MM10 S2 gRNA. RT-LAMP was performed at 65 °C for 15 minutes under optimized conditions, which encompassed doubling the concentration of B3 to 0.4 µM, using both full-length and 1nt-truncated internal primers (1.6 µM each), including the swarm primers (1.6 µM each), and adding 0.15U Q5 polymerase and 0.1 M glycine into each reaction. Fluorescence readings using a microplate reader after 10 min of cleavage reaction at 37 °C are shown. Data represent mean ± s.e.m. (n = 5 [2E6] or 6 [other copy numbers] biological replicates). f Analytical LoD for enAsCas12a when a S254F mutation was present in the viral template. The nuclease was assembled either with the S2 gRNA alone or with both the S2 and S6 gRNAs. These gRNAs were designed to be perfect matched against the reference SARS-CoV-2 genome. RT-LAMP was performed at 65 °C for 15 min under optimized conditions. Fluorescence readings after 10 min of cleavage reaction at 37 °C are shown. Data represent mean ± s.e.m. (n = 5 biological replicates). g Similar experiments to f, except that a different reporter was used and a dipstick was added to each sample tube after 10 minutes of cleavage reaction. Bands appeared on the dipsticks by 2 min. The red arrow indicates the test bands, while the green arrow indicates the control bands. Ratios of test band intensity to control band intensity are given under each dipstick. Source data are available in the Source Data file.