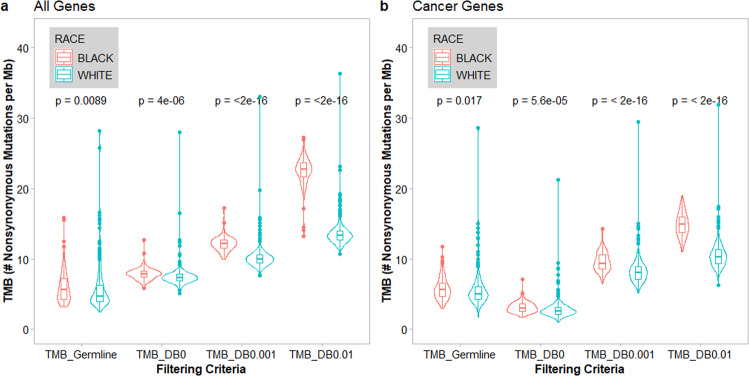
Fig. 1. Impact of variant filtering criteria on TMB calculation.
The TMB values were calculated as number of nonsynonymous mutations per Mb of coding regions. Four criteria were applied to identify patient-specific somatic mutations: (1) TMB_Germline: excluding variants in patient-matched germline exome; (2) TMB_DB0: excluding all variants reported by 1000G or ExAC; (3) TMB_DB0.001: excluding variants with MAF ≥ 0.1% in 1000G or ExAC; and (4) TMB_DB0.01: excluding variants with minor allele frequency (MAF) ≥ 1% in 1000G or ExAC DBs. The violin and box plots in red are TMB values from Black patients, and blue are from White Patients. a Comparisons of TMBs calculated from all protein-coding genes in Black and White patients from four variant filtering criteria. b Comparisons of TMB calculated from 1059 cancer genes in Black and White patients from four variant filtering criteria.