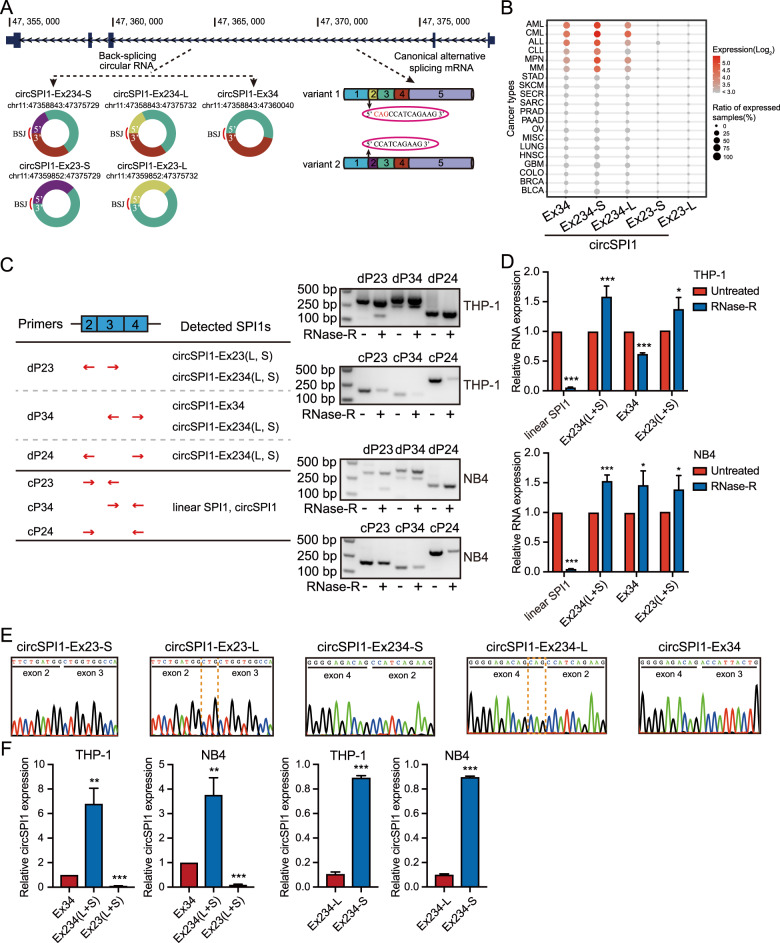
Fig. 1. Identification and characterization of circSPI1 in AML.
A. Schematic overview showed circSPI1 isoforms in SPI1 locus. Five circSPI1s were retrieved from the circRNA compendium MiOncoCirc database. BSJ back-splicing junction. B CircSPI1s were hematopoietic lineage-specific circRNAs. The expression of five circSPI1 isoforms was analyzed in diverse cancer types from MiOncoCirc database. AML acute myeloid leukemia, CML chronic myeloid leukemia, ALL acute lymphocytic leukemia, CLL chronic lymphocytic leukemia, MPN myeloproliferative neoplasm, MM multiple myeloma, STAD stomach adenocarcinoma, SKCM skin cutaneous melanoma, SECR secretory cancer, SARC sarcoma, PRAD prostate adenocarcinoma, PAAD pancreatic adenocarcinoma, OV ovarian serous cystadenocarcinoma, MISC miscellaneous cancer, LUNG lung adenocarcinoma, HNSC head and neck squamous cell carcinoma, GBM glioblastoma multiforme, COLO colon adenocarcinoma, BRCA breast invasive carcinoma, BLCA bladder urothelial carcinoma. C The existence of circSPI1s was validated with divergent and convergent primers in THP-1 and NB4 cells. The schematic diagram illustrated the location of divergent and convergent primers and the recognized circSPI1s of divergent primers (left panel), then circSPI1s were detected by RT-PCR (right panels) with divergent and convergent primers before and after treatment with RNase R. Two major bands produced by the divergent primer pair dP23 corresponded to circSPI1-Ex234 (L and S) and circSPI1-Ex23 (L and S), respectively. Two major bands produced by the divergent primer pair dP34 corresponded to circSPI1-Ex234 (L and S) and circSPI1-Ex34. One band produced by dP24 corresponded to circSPI1-Ex234 (L and S). Convergent primers were served as a negative control. dP divergent primers, cP convergent primers. D CircSPI1 isoforms were analyzed by RT-qPCR in THP-1 and NB4 cells. E The junction sites of circSPI1 isoforms were identified by sanger sequencing. All detected amplicons by divergent primers were subjected to TA clone, and then positive clones were performed sanger sequencing. F The relative expression levels of circSPI1 isoforms were analyzed in THP-1 and NB4 cells. The left two panels: RT-qPCR analysis measured the relative levels of five circSPI1s. The right two panels: TaqMan probe-based RT-qPCR assays determined the expression levels of circSPI1-Ex234-L and circSPI1-Ex234-S. All bar graphs represent the average of three independent replicates and error bars are SD, *P < 0.05, **P < 0.01, ***P < 0.001.