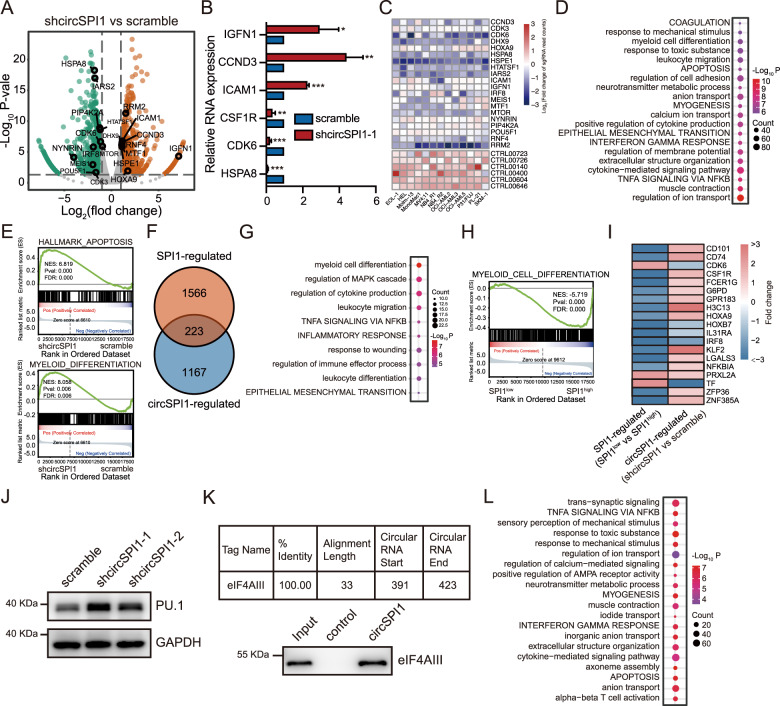
Fig. 4. CircSPI1 antagonizes SPI1 to modulate myeloid differentiation.
A Volcano plot of differentially expressed genes after circSPI1 knockdown. The vertical lines correspond to 2.0-fold up and down, and the horizontal line represents a P value of 0.05. B The expression changes of representative DEGs after circSPI1 knockdown were validated by RT-qPCR. C Representative DEGs upon circSPI1 knockdown were essential for the survival of AML cells. Normalized read counts of sgRNA in AML cells transfected with the CRISPR-cas9/sgRNA library before and after population doublings were showed. D Functional enrichment analysis showing significant GO terms and hallmark gene sets among circSPI1-regulated genes. E GSEA showing that differentiation and apoptosis were enriched using shcircSPI1-regulated genes. F The overlap of regulated genes between shcircSPI1 and shSPI1. G Top ten enriched functional terms for shared genes between circSPI1 and SPI1. H Differentiation was enriched using SPI1-regulated genes. I Heatmap showing that myeloid differentiation-associated genes were oppositely regulated by circSPI1 and SPI1. J The protein level of SPI1 was detected with or without circSPI1 knockdown. GAPDH served as the loading control. The original gels and three replicates are presented in Supplementary Fig. 3A. K CircSPI1 interacted with eIF4AIII in AML cells. The binding potential of circSPI1 with eIF4AIII was predicted by the Interactome database (upper panel). The interaction between circSPI1 and eIF4AIII was detected by western blotting combined with RNA pulldown (lower panel). Total lysates prepared from NB4 cells were subjected to pulldown with circSPI1 and negative control probes, followed by western blotting. The original gels and three replicates are presented in Supplementary Fig. 3B. L Functional enrichment of genes regulated by circSPI1 but not SPI1 was summarized using annotated GO terms and hallmark gene sets. All bar graphs represent the average of three independent replicates and error bars are SD, *P < 0.05, **P < 0.01, ***P < 0.001.