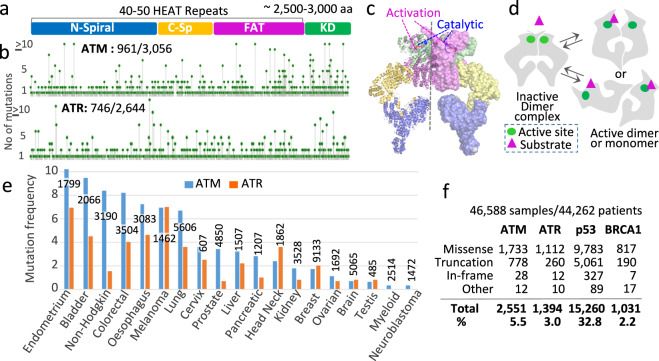
Fig. 1. ATM/ATR mutations in human cancer.
a ATM/ATR proteins are large (~2200–3000 residues) and comprise 40–50 tandem HEAT repeats followed by a highly conserved kinase domain45. The HEAT repeat domain is divided into three conserved structural motifs referred to as the N-spiral/solenoid, C-spiral/bridge/pincer, and FAT domains9–12. b Screen shot images from the cBio Cancer Genomics Portal website (http://cbioportal.org). The images show the location and frequencies of ATM/ATR residues mutated in 46,588 tumor samples in the database. In ATM, 961 of the 3056 residues are mutated; in ATR, 746 of the 2644 residues are mutated (Supplementary Data 1 and 2). c A cryo-EM model structure of the ATM enzyme complex (PDB 5NPO, 5.70 Å)9. The complex is a dimer comprising two identical protomers that share multiple interfaces. The protomer on the right and left are in surface and cartoon representations, respectively. Both protomers are shown in four colors, each corresponding to the indicated domain in a. The catalytic and activation loops in the active site are shown in red and blue, respectively. d Model: the ATM/ATR and Tel1/Mec1 enzyme complexes are in dynamic equilibrium between an inactive and active conformations. Under unchallenged conditions the dimeric complex exists as a minimally active enzyme, in which substrate accessibility to the active site is sterically hindered. Stress-dependent conformational change activates the enzyme by relieving the block. The activation may take place in the dimer context or entail a dimer-to-monomer transition9–12,46. e ATM/ATR mutation frequencies in the indicated cancer types (Supplementary Table 1; http://cbioportal.org). The number of tumor samples analyzed for each cancer type is as indicated. f The number of cancer-associated missense, truncation, in-frame, or other mutations in ATM, ATR, P53, or BRCA1 (http://cbioportal.org).