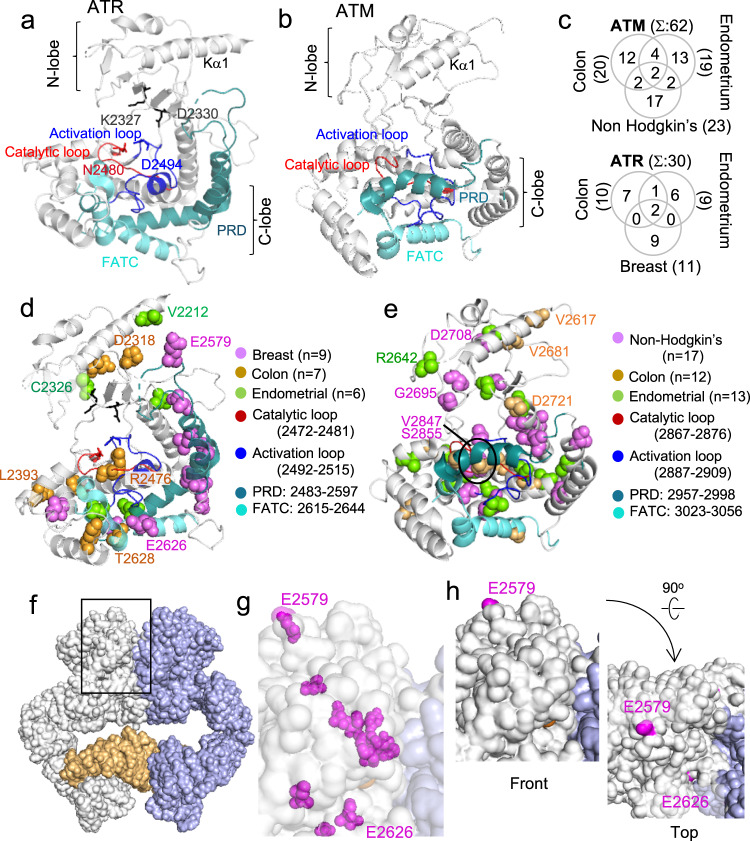
Fig. 4. Molecular modeling analysis of ATM/ATR kinase domain residues implicated in cancer.
a Kinase domain of an active ATR enzyme complex (PDB 5YZO, 4.70 Å)10. The PRD is positioned away from catalytic center to allow substrate access. The K2327 and D2330 support ATP association. The N2480 and D2494 stabilizes Mg2+ for catalysis10. b Kinase domain of an inactive ATM enzyme complex (PDB 5NPO, 5.70 Å)9. The PRD sterically hinders substrate access to the catalytic center. c Extent of overlap among the ATM/ATR residues mutated in the indicated cancer type that are the kinase domain (Fig. 2b, Supplementary Fig. 2a, and Supplementary Data 3 and 4). d, e The kinase domains of ATR (d) and ATM (e), showing location of the conserved residues mutated in the indicated cancer type. Labeled residues are solvent accessible (h, g) Supplementary Fig. 4f, g). f A surface representation of the ATR enzyme complex (PDB 5YZO, 4.70 Å)10. The left and right ATR polypeptides are white and blue, respectively, and Ddc2 is in bright orange. Black rectangle: the kinase domain regions highlighted in g and h. g Higher resolution image of the area highlighted by a black rectangle in f. Magenta spheres: residues mutated in breast cancer shown in d. The transparency setting was set to “on” to visualize both the buried and exposed residues. Only the E2579 and E2626 are exposed (h). h Same as in g except that the transparency setting was “off”. Front and top views show that only the E2579 and E2626 are solvent accessible.