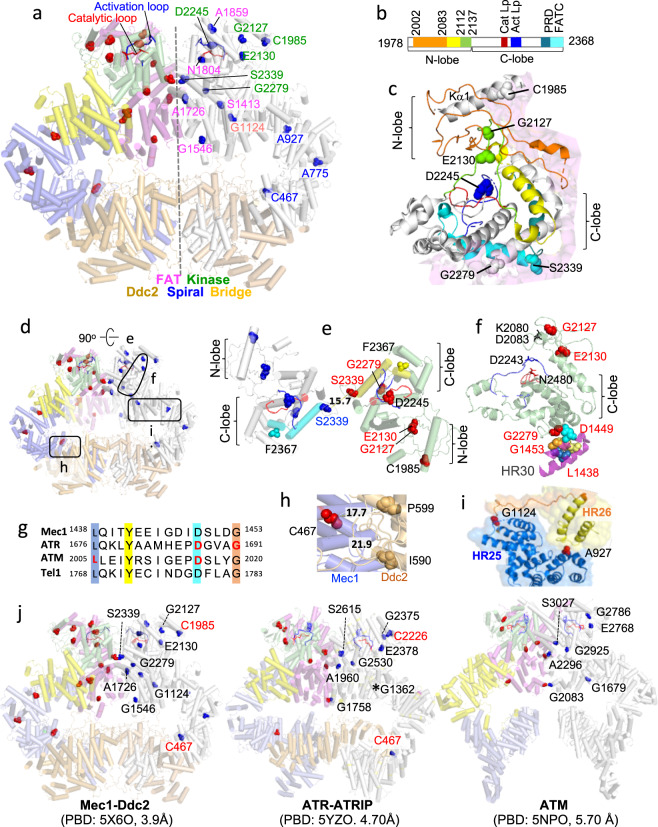
Fig. 6. Molecular modeling analysis of Mec1 residues involved in the DDR.
a The 15 critical Mec1 residues are mapped onto a cryo-EM model of the inactive dimeric Mec1–Ddc2 enzyme complex (PDB 5X6O, 3.9 Å)11. The dashed black line in the middle denotes the twofold symmetry axis. The complex is in cylindrical helices representation. The protomer on the left-hand side is in blue, yellow, violet, and green, representing the spiral, bridge, FAT, and kinase domains, respectively. The protomer on the right-hand side is in white. Both Ddc2 chains are shown in light brown. The 15 residues in the left and right protomers are shown in red and blue sphere representation. b Schematic representation of the Mec1 kinase domain, which comprises the N- and C-lobes. The N-lobe contains two extended loops shown in orange (residues 2002–2083) and green (2112–2137). The first loop is preceded by the kα1 (1978–2002) shown in white (c). Between the two loops is another helical (2083–2112 shown) shown in yellow (c). The C-lobe contains the critical activation and catalytic loops, and several regulatory motifs including the PRD and FATC. c A round helices representation of the Mec1 kinase domain. The six residues identified in our screen are shown in sphere representation in the colors corresponding to those shown in b. The neighboring FAT domain is shown in faint pink. d Lower resolution image of the model in a, showing the areas that are highlighted in e, f, h, and i. e Top view of the dimeric enzyme complex in cylindrical helices representation depicting proximity of the two S2339 residues; only the two kinase domains are shown. The kinase domain on the left is in white except the FATC, which is in cyan. The kinase domain on the right is in pale green except the FATC, which is in yellow. The activation and catalytic loops are shown in blue and red, respectively. The six mutated residues are shown in blue (left) or red (right). The two S2339 residues are separated by 15.7 Å. Red label: the corresponding ATM and/or ATR residue is mutated in cancer. Cyan and yellow: FATC. F2367: N-terminus of the Mec1 polypeptide in the model; the last residue, W2368, is missing in the published Mec1–Ddc2 complex11. f The G2279 is at an interface between the kinase domain and the HR30 of the FAT domain. The HR30 is ~800 residues away from the G2279 and contains four residues that are conserved in ATR, ATM, and Tel1 (g). Red residue: corresponds to an ATM and/or ATR residue mutated in cancer. The K2080 and D2083 correspond to the ATR K2327 and D2330 involved in ATP association, respectively. The N2229 and D2243 correspond to the ATR N2480 and D2494 that stabilize Mg2+ for catalysis, respectively10 (Fig. 4a). The G2127 and E2130 are two other kinase domain residues identified in the current study (a, c). g Sequence alignment of the Mec1 HR30 and the corresponding HRs in ATR, ATM, and Tel1. The four shaded residues are conserved across the four proteins. Color of the shade correspond to the color of each residue in f. The ATM L2005 and D2016, and the ATR D1687 and G1691 are mutated in cancer (Supplementary Fig. 7b, and Supplementary Data 3, 4, and 6). h The Mec1 C467 is at the interface between the Mec1 spiral domain (blue) and Ddc2 (light brown). The C467 is ~20 Å from the Ddc2 I599 and P599, which correspond to the ATRIP L662 and P671 mutated in cancer, respectively (http://cbioportal.org). i The Mec1 A927 in the spiral domain and the G1124 is in the linker (1122–1148)11 are found at the interface between the spiral (blue) and bridge (yellow) domains. j Cryo-EM models Mec1, ATR, and ATM enzyme complexes showing location of the conserved residues critical for resistance to HU/MMS in yeast (Fig. 5a, c). The seven black residues are conserved in Mec1, ATR, and ATM. The two red residues are conserved only in Mec1 and ATR. *ATR G1362 is in a flexible region (Supplementary Fig. 7d) and missing in the published model10.