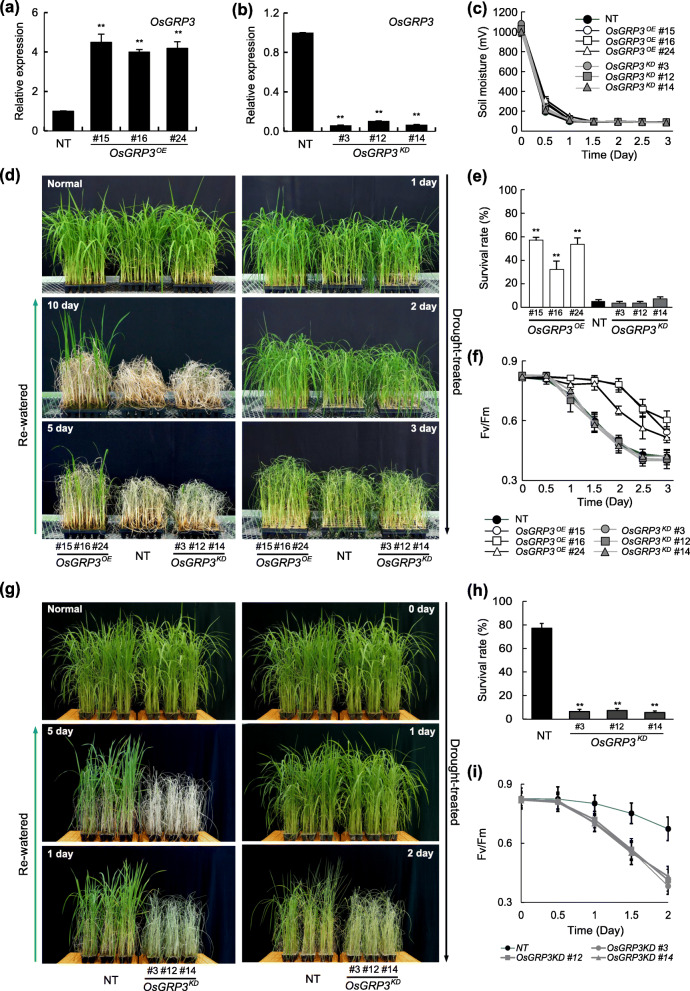
Fig. 2.
Overexpression of OsGRP3 enhances drought tolerance in rice. Relative expression levels of OsGRP3 in OsGRP3 overexpressing (OsGRP3OE) (a) and RNAi-mediated OsGRP3 suppressing (OsGRP3KD) (b) transgenic plants. Total RNAs were extracted from leaves of two-week-old rice seedlings and used for qRT-PCR analysis. Rice UBIQUITIN (OsUBI) was used as an internal control for normalization. a and b Data represent the mean value ± standard deviation (SD) (n = 3, biological replicates). c Soil moisture content was monitored during drought treatment. Data represent the mean value ± standard deviation (SD) of 30 independent measurements performed at different locations of pots. d The drought-induced symptoms of non-transgenic (NT), OsGRP3OE and OsGRP3KD transgenic plants during drought stress and re-watering were visualized by taking pictures at indicated time points. e The survival rate of NT, OsGRP3OE and OsGRP3KD transgenic plants was calculated by counting the number of plants recovered from drought stress after re-watering. f Photochemical efficiency was analyzed by measuring the leaf chlorophyll fluorescence (Fv/Fm) of non-transgenic (NT), OsGRP3OE and OsGRP3KD transgenic plants during drought treatments. g The drought-induced symptoms of NT, OsGRP3OE and OsGRP3KD transgenic plants during drought stress and re-watering were visualized by taking pictures at indicated time points. h The survival rate of NT, OsGRP3OE and OsGRP3KD transgenic plants was calculated by counting the number of plants recovered from drought stress after re-watering. i Fv/Fm value of NT and OsGRP3KD transgenic plants during drought treatments. Value for each time point represents the mean ± standard deviation (SD) (n = 30, number of plants). Asterisks indicate a statistically significant difference compared with NT. **P < 0.01; One-way ANOVA