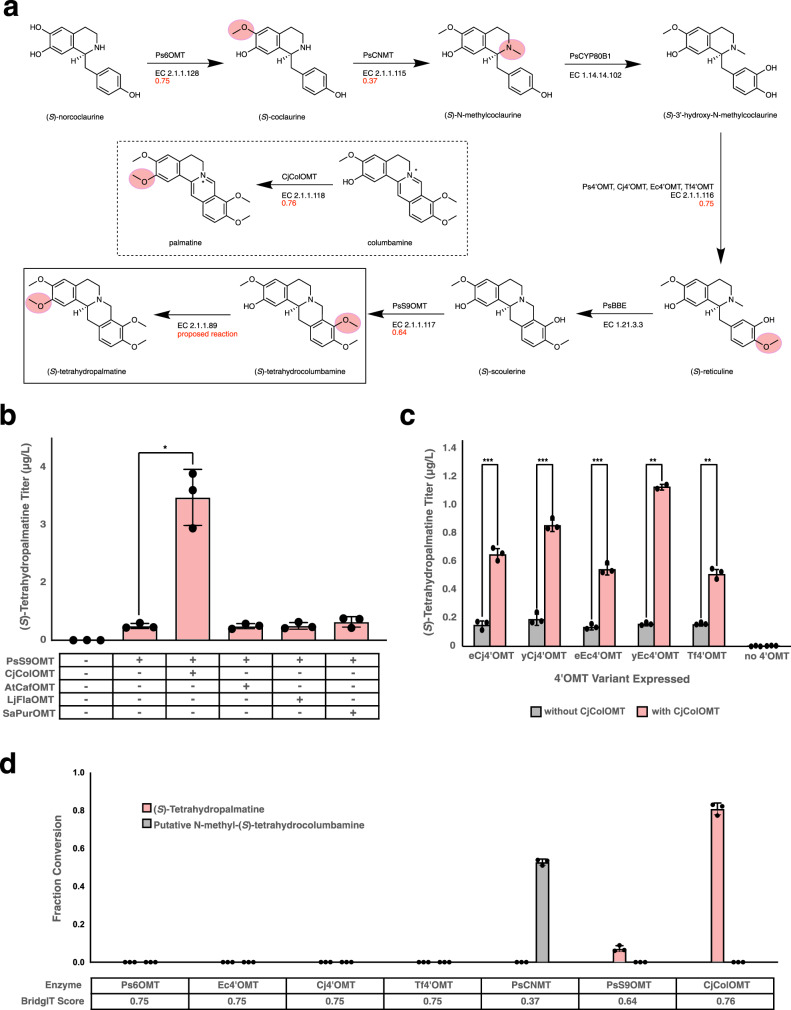
Fig. 3. In vivo and in vitro activity of predicted enzymes.
a Biosynthetic pathway from (S)-norcoclaurine, the first dedicated intermediate in the pathway, to (S)-tetrahydropalmatine. The specific enzyme(s) used in our strains are indicated above each reaction arrow, while below each arrow is the enzyme class and, for methyltransferases, the BridgIT score (in red) obtained for the likelihood of members of that class to perform our proposed reaction. Our proposed reaction, the methylation of (S)-tetrahydrocolumbamine to afford (S)-tetrahydropalmatine, is shown in the box at the bottom left. Shown in dotted lines is the native reaction of CjColOMT, the enzyme which was predicted and demonstrated to perform our proposed reaction. The site of methylation of each methyltransferase is highlighted on its product in pink. b De novo production of (S)-tetrahydropalmatine in yeast strains engineered to express members of the two most downstream O-methyltransferase classes (S9OMT & ColOMT) predicted by BNICE.ch & BridgIT to accept (S)-tetrahydrocolumbamine as a substrate. PsS9OMT is integrated into the yeast genome, while CjColOMT, AtCafOMT, LjFlaOMT, and SaPurOMT were expressed from a high-copy plasmid; the first two strains shown contain an empty version of this plasmid. Strains were cultured in selective media (YNB-Ura) with 2% dextrose, 2 mM L-DOPA, and 10 mM ascorbic acid at 30 °C for 120 h before LC-MS/MS analysis of the growth media. Data are presented as mean values ± the standard deviation of three biologically independent samples. Asterisks represent Student’s two-tailed t-test: *p < 0.05, **p < 0.01, ***p < 0.001. Exact p-values are given in Supplementary Table 8. c In vitro reactions of purified methyltransferases on (S)-tetrahydrocolumbamine to produce (S)-tetrahydropalmatine (shown in pink) or the putative N-methyl-(S)-tetrahydrocolumbamine product (shown in gray). BridgIT score denotes the score obtained by BridgIT for the enzyme class to which each enzyme belongs. Data are presented as mean values ± the standard deviation of three biologically independent samples. Asterisks represent Student’s two-tailed t-test: *p < 0.05, **p < 0.01, ***p < 0.001. Exact p-values are given in Supplementary Table 8. d De novo production of (S)-tetrahydropalmatine in yeast strains engineered to express alternative 4’OMTs. Strains were cultured in selective media (YNB-Ura) with 2% dextrose, 2 mM L-DOPA, and 10 mM ascorbic acid at 30 °C for 72 h before LC-MS/MS analysis. Data are presented as mean values ± the standard deviation of three biologically independent samples. Source data underlying Fig. 3b–d are provided as a Source Data file.