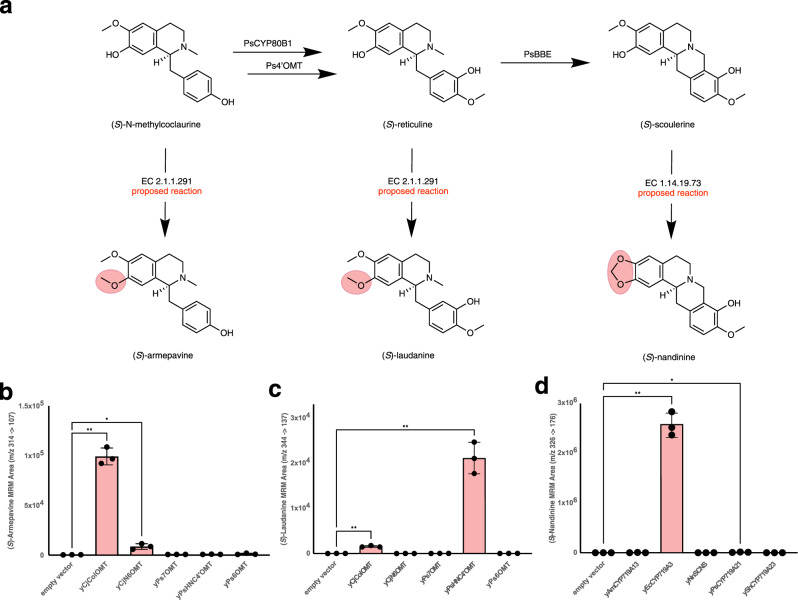
Fig. 4. Pathway expansion to produce three additional derivatives de novo.
a Portion of noscapine biosynthetic pathway from (S)-N-methylcoclaurine to (S)-scoulerine with potential additional enzymatic steps to produce three derivatives: (S)-armepavine, (S)-laudanine, and (S)-nandinine. For steps upstream of (S)-N-methylcoclauine, downstream of (S)-scoulerine, or the structure of the omitted intermediate ((S)-3′-hydroxy-N-methylcoclaurine), see Fig. 3a. De novo production of the three derivatives are shown in panels b ((S)-armepavine production), c ((S)-laudanine production), and d ((S)-nandinine production). The y-axes of each graph in panels b–d show the integrated area of the peak measured by LC-MS/MS multiple reaction monitoring (MRM) at the quantifier transition indicated for each compound (see Supplementary Table 7 for additional details). For each derivative, all genes necessary for biosynthesis of the substrate are integrated into the genome of the parent strain (CSY1322 for (S)-armepavine production; CSY1171 for (S)-laudanine production; CSY1320 for (S)-nandinine production), while the genes encoding the enzymes predicted by BridgIT (shown on the x-axes of the graphs in panels b–d) are expressed from a high-copy plasmid. Strains were cultured in selective media (YNB-Ura) with 2% dextrose, 2 mM L-DOPA, and 10 mM ascorbic acid at 25 °C for 96 h before LC-MS/MS analysis of the growth media. Data are presented as mean values ± the standard deviation of three biologically independent samples. Asterisks represent Student’s two-tailed t-test: *p < 0.05, **p < 0.01, ***p < 0.001. Exact p-values are given in Supplementary Table 8. Source data underlying Fig. 4b–d are provided as a Source Data file.