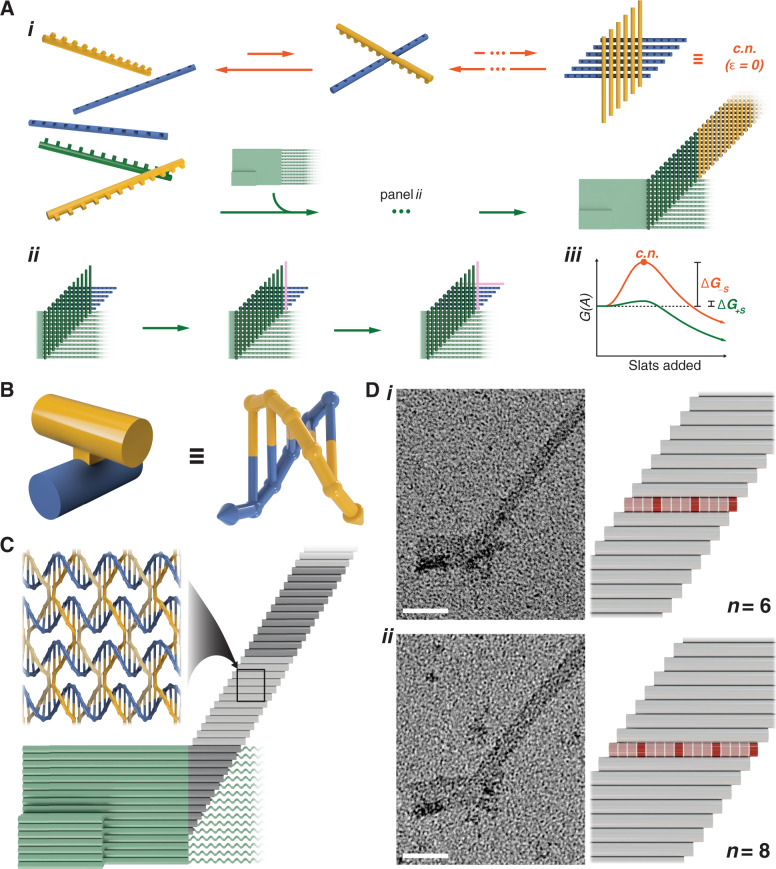
Fig. 2. Crisscross polymerization as implemented with ssDNA slats.
Ai Ribbon assembly of abstract v6 x-slats (blue) and y-slats (gold). Upper pathway shows transient association of free slats, where the critical nucleus (c.n.) requires a multitude of unfavorable slat additions. Lower pathway uses a seed (light green) to capture twelve nuc-y-slats (dark green) along with six x-slats. Aii, Subsequent slats (magenta) are recruited in alternating perpendicular fashion for ribbon growth. Aiii, Qualitative energy landscape for unseeded (orange) versus seeded (green) nucleation and growth of slats using irreversible growth conditions. B Each binding domain in the abstract model is a half turn of DNA. C Crisscross polymerization of ssDNA slats (blue x- and gold y-slats) as triggered by a DNA-origami seed (light green). Straight cylinders represent double helices. D Left are negative-stain TEM images of v6 (n = 6) and v8 (n = 8) ribbons, in (i) and (ii), respectively. Right models show width differences, where each boxed cell is a single half-turn domain. Light-red or dark-red cells correspond to either five or six base-pair domains. Scale bars are 50 nm.