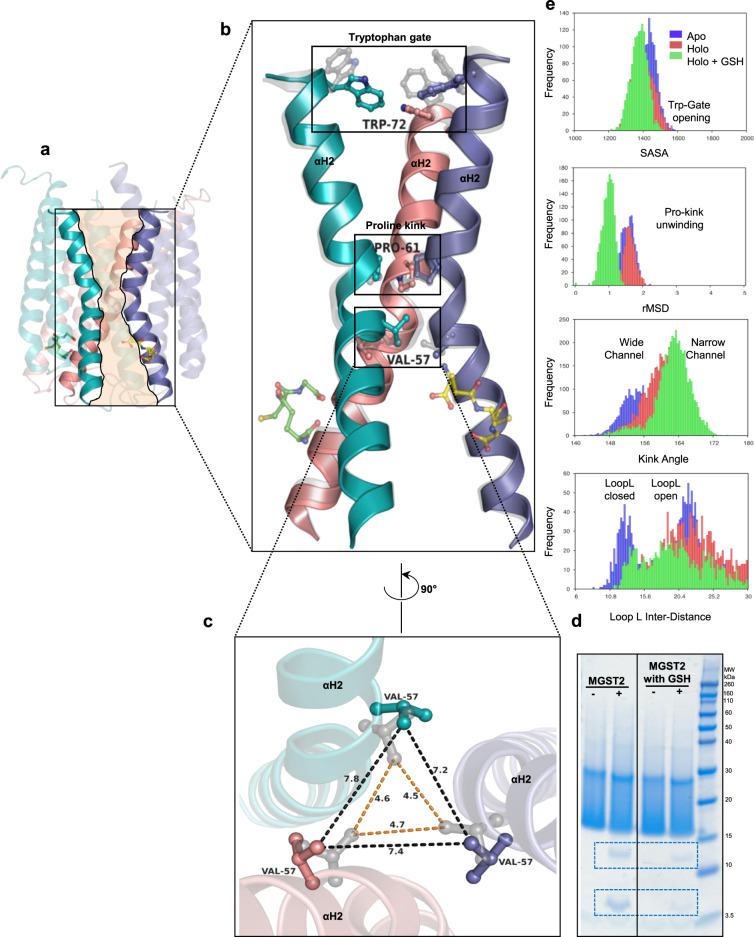
Fig. 6. Comparison of apo- and holo-MGST2 structures unveils conformational changes of the central channel associated with co-substrate binding.
a Holo-MGST2 (cartoon) with highlighted central channel (beige) created by αH2 of each monomer, which connects the cytoplasmic and luminal part of the enzyme. b Superimposed αH2 of holo-MGST2 (opaque cartoon) and apo-MGST2 (gray translucent cartoon) displaying dynamic residues Trp72, Pro61, and Val57 located at the trimer interface. Bound GSH molecules are depicted in ball and stick representation. c Widening of the cytoplasmic cone represented by a triangle between adjacent c∂-distances of Val57 in apo (gray translucent ball and stick), which increases from ~4.7 Å (orange dashed lines) to ~7.8 Å (blue dashed lines) in holo-MGST2 (opaque ball and stick). d Trypsin digestion of MGST2 enzyme in the presence of GSH tends to be more stable as reflected in less intensity of trypsin-digested bands compared with the enzyme digested with no substrates. Similar band pattern was observed in three independent digestions. Source data are provided as a Source data file. e Key dynamic descriptors along MGST2 axis in MD simulations: the Trp-gate is closed in the presence of GSH making the luminal cavity less accessible for solvent (top); pro-kink unwinding is observed in the absence of GSH as indicated by the shift in rMSD (second top); kink angle is reduced by GSH binding making the central pore narrower (third top); loop L has a marked open/close dynamics in the apo state (bottom).