Fig. 7.
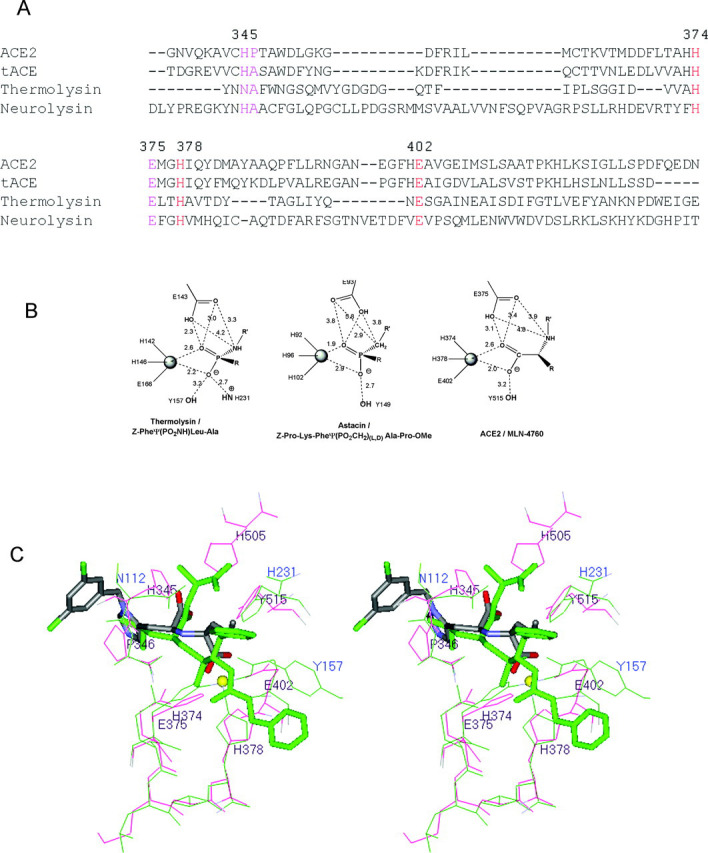
Structural homology for the ACE2 catalytic motif to other members of the HEXXH metallopeptidase clans.A, shown is the structure-based sequence alignment of ACE2, tACE, thermolysin, and neurolysin. The conserved residues correspond to the catalytic motif for these enzymes (colored red (zinc binding) and magenta). Sequence numbering is for ACE2. B, the catalytic motifs for thermolysin and astacin bound to transition state analogs Z-Pheψ(PO2NH)-Leu-Ala and Z-Pro-Lys-Pheψ(PO2CH2)-dl-Ala-Pro-OMe, respectively (30, 42), are compared with the ACE2 complex with MLN-4760. Distances are measured in angstroms. C, shown is the superposition of the catalytic motifs of ACE2 (red) and thermolysin (green). Eight α-carbon atoms corresponding to residues 345, 346, 374-378, and 402 of MLN-4760 bound ACE2 were superimposed onto the equivalent α-carbon atoms of Z-Pro-Lys-Pheψ(PO2CH2)-dl-Ala-Pro-OMe-bound thermolysin (see sequence alignment in A) with an r.m.s. deviation of 0.49 Å. Bound inhibitors are shown in stick rendering with default atom coloring for MLN-4760 and green coloring for Z-Pro-Lys-Pheψ(PO2CH2)-dl-Ala-Pro-OMe. The zinc ion is shown as a yellow sphere. ACE2 labels are black, and thermolysin labels are blue. ψ indicates replacement of the peptide bond by the group in parentheses.
