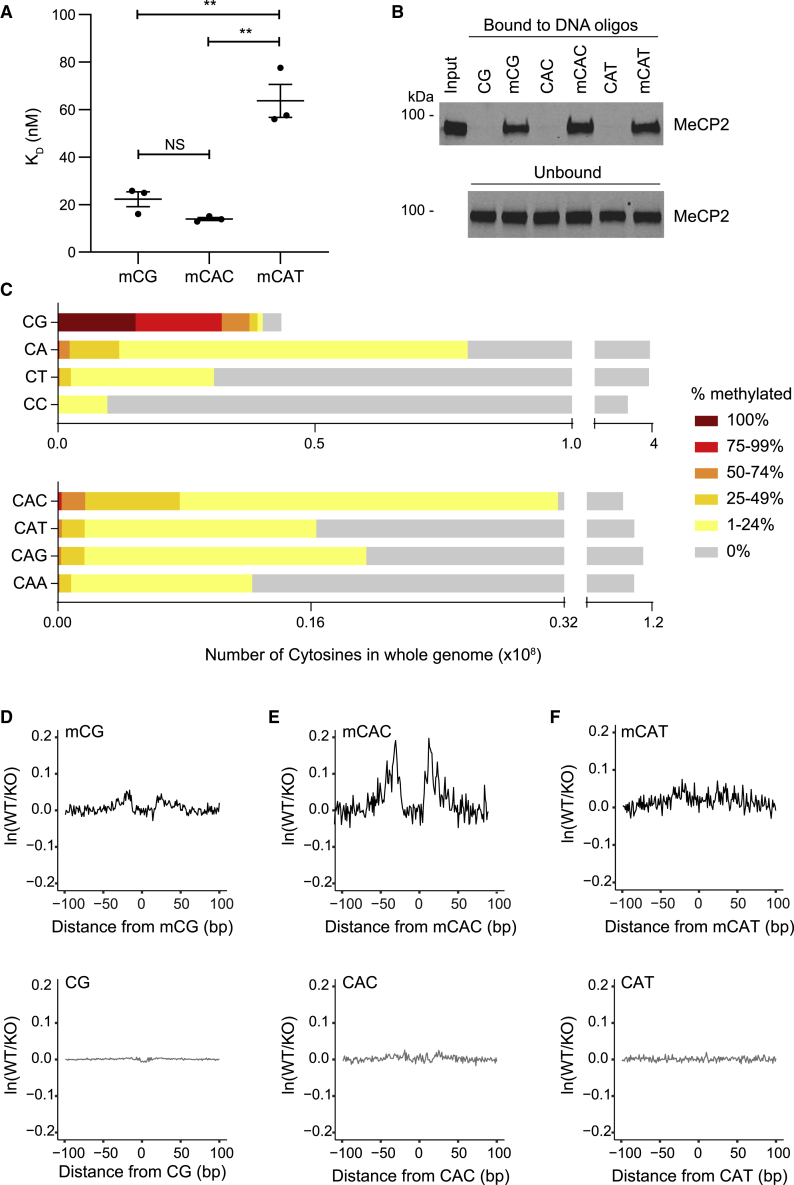
Figure 1.
MeCP2 binds mCG and mCAC, and to a lesser extent, mCAT
(A) Bio-layer interferometry (BLI) analysis of the interaction between the MBD of MeCP2 and methylated DNA probes containing a single mCG, mCAC, or mCAT site. KD values: mCG 22.25 ± 3.12 nM; mCAC 13.90 ± 0.64 nM; mCAT 63.71 ± 6.94 nM (means ± SEMs). Binding affinity to mCG and mCAC is similar (not significant [NS] p = 0.06); binding to mCAT is significantly weaker than mCG and mCAC (∗∗p = 0.006 and ∗∗p = 0.002, respectively), t tests.
(B) Western blot analysis of MeCP2 following pull-down from rat brain nuclear extracts using immobilized DNA containing unmethylated and methylated CG, CAC, and CAT (top) and unbound MeCP2 in the supernatant (bottom).
(C) Whole-genome bisulfite sequencing analysis in the mouse hypothalamus (Lagger et al., 2017) showing the distribution of methylation at each CN dinucleotide (top) and each CAN trinucleotide (bottom). Sites were binned by level of methylation.
(D–F) ATAC-seq footprinting analysis of MeCP2 over methylated (top) and unmethylated (bottom) CG (D), CAC (E), and CAT (F) sites in the mouse hypothalamus.