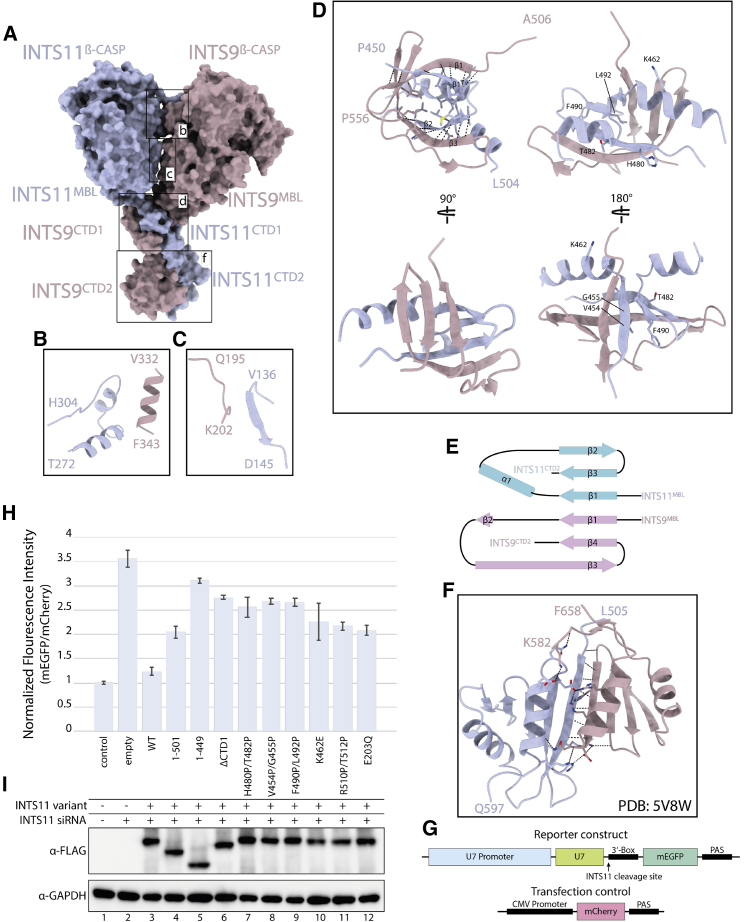
Figure 3.
Structural basis of INTS9/11 heterodimerization
(A) Surface representation of the model, showing contact areas. Interfaces involved in dimer formation are highlighted with close ups.
(B and C) Insets showing the proximity of the two nuclease domains; no polar contacts could be identified between these regions.
(D) A β-barrel-like structure formed by the CTD1 domains of INTS9 and INTS11, highlighting intermolecular β-sheets formed by the two proteins.
(E) A topology plot of the CTD1 dimer, showing the intermolecular β-sheet.
(F) CTD2 dimer of INTS9 and INTS11, as reported previously (PDB: 5V8W).
(G) Schematics of the constructs used in the GFP-based U7 snRNA in vivo processing assay.
(H) mEGFP/mCherry fluorescence intensity readout from the depletion/rescue experiment assessing the functionality of different INTS11 variants. “Empty” refers to the condition where no protein was overexpressed. Error bars represent standard deviation from 3 individual measurements.
(I) Western blot showing protein expression levels of the transgenes used in the depletion/rescue experiment (H).