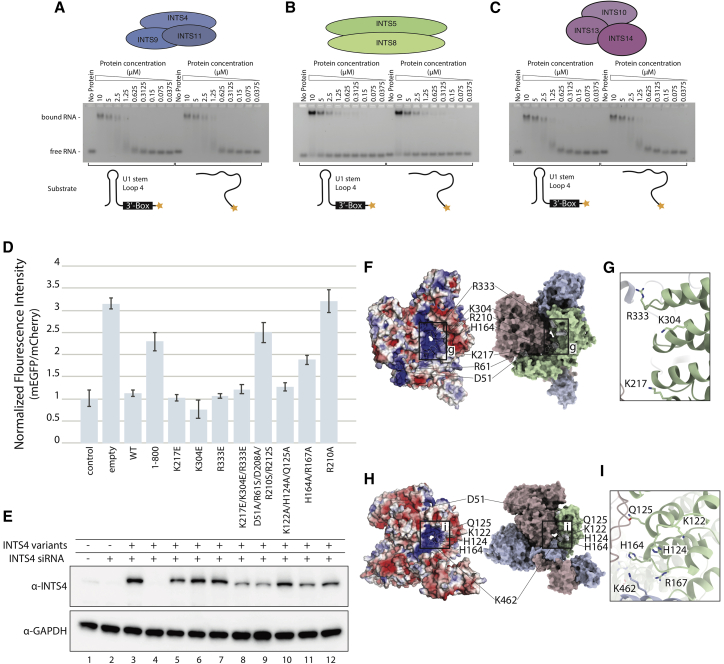
Figure 6.
Electropositive surface of INTS4/9/11 and RNA-binding properties of different Integrator sub-complexes
(A–C) EMSA titration assay showing the weak RNA-binding properties of the INTS4/9/11 complex toward a 3′-box-containing U1 pre-snRNA substrate (left panel) and scrambled RNA sequence of the same length (right panel). The same EMSA assay was performed for INTS5/8 (B) and INTS10/13/14 (C).
(D) mEGFP/mCherry fluorescence intensity readout from the depletion/reconstitution experiment assessing functionality of INTS4 variants designed to disrupt the putative RNA-binding surface. “Empty” refers to the condition where no protein was overexpressed. Error bars represent standard deviation from 3 individual measurements.
(E) Western blot showing protein expression levels of the INTS4 variants used in the depletion/reconstitution experiment (D). The lack of signal for INTS4 in lane 4 was expected because the antibody used for detection was raised against the peptide comprising residues 913–963.
(F and H) Surface electrostatic potential calculated with Adaptive Poisson-Boltzmann Solver (APBS) of the INTS4/9/11 complex, showing the location of the point mutations tested with the reporter system.
(G and I) Magnified view of the structural environment of the discussed residues.