Table 1.
Estimated docking scores (in kcal/mol) and binding features for rutin and the top nine potent analogs against SARS-CoV-2 Mpro.
| No. | PubChem code | 2D-Chemical Structure | Docking Score (kcal/mol) | Cluster Populationa | Binding Featuresb |
|---|---|---|---|---|---|
| 1 | Rutin (PubChem-528-080-5) | 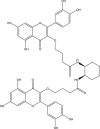 |
−7.2 | 43 | ASP187 (1.90 Å), HIS164 (2.25 Å), GLU166 (2.05 Å), GLN192 (2.86 Å), THR190 (2.19, 1.92 Å) |
| 2 | PubChem-137-399-195 | 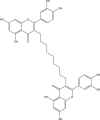 |
−10.9 | 38 | GLN189 (2.22 Å), GLU166 (1.92, 1.90, 2.99 Å), GLY143 (1.95, 2.88 Å), LEU141 (2.57 Å), SER144 (1.83 Å) |
| 3 | PubChem-129-716-607 | 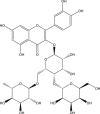 |
−10.7 | 37 | THR26 (2.07 Å), GLU166 (2.06 Å), HIS164 (2.03 Å), TYR54 (2.33 Å), ASP187 (2.32 Å), THR190 (2.20, 2.02 Å) |
| 4 | PubChem-893-333-51 | 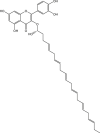 |
−9.9 | 47 | THR190 (1.92 Å), ASP187 (1.90 Å), GLU166 (2.02, 1.85, 1.78 Å), HIS164 (2.28 Å), HIS41 (2.40 Å), LEU141 (1.83 Å), SER144 (1.96 Å), GLY143 (2.37 Å), CYS145 (2.59, 2.61 Å) |
| 5 | PubChem-123-170-316 | 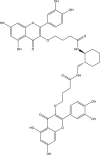 |
−9.9 | 27 | THR190 (1.85 Å), GLU166 (1.94 Å), HIS164 (2.45 Å), ASP187 (1.93 Å) |
| 6 | PubChem-142-513-769 | 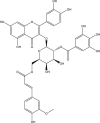 |
−9.8 | 29 | GLY143 (1.62 Å), ASN142 (2.33 Å), PHE140 (2.09 Å), HIS163 (2.08 Å), HIS164 (2.74 Å), GLU166 (2.13, 2.16, 2.96 Å), PRO168 (2.05 Å) |
| 7 | PubChem-101-020-740 | 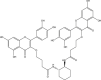 |
−9.7 | 35 | THR190 (1.98, 2.08 Å), GLN189 (1.99 Å), GLU166 (1.93 Å), ASP187 (2.78 Å), HIS164 (2.92 Å), HIS163 (2.09 Å), SER144 (2.12 Å), LEU141 (2.21 Å), LEU141 (1.98, 2.21 Å) |
| 8 | PubChem-142-513-754 | 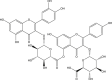 |
−9.7 | 37 | LEU141 (2.79 Å), HIS163 (1.65 Å), GLU166 (2.48, 2.16 Å), ASP187 (2.00 Å), THR190 (2.18, 2.26 Å) |
| 9 | PubChem-885-071-27 | 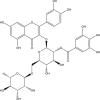 |
−9.5 | 42 | THR26 (2.07 Å), ASN142 (2.06 Å), PHE140 (2.8 Å), HIS163 (1.99 Å), GLU166 (2.07 Å), MET165 (2.4, 2.72 Å), GLN192 (2.73 Å), THR190 (2.21, 1.73 Å), PRO168 (2.20, 1.89 Å) |
| 10 | PubChem-101-133-681 |
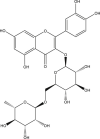 1 1 |
−9.2 | 34 | GLY143 (2.08 Å), GLU166 (1.70, 2.20, 1.86 Å), THR190 (2.22, 2.54 Å) |
Number of conformations in the largest cluster.
Only hydrogen bonds (in Å) were listed.
