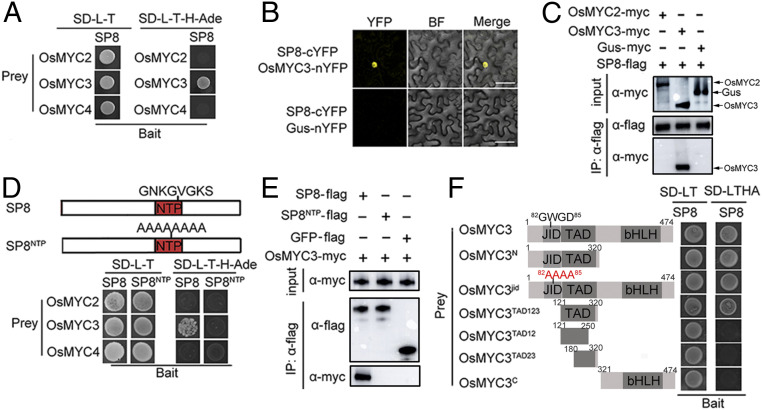
Fig. 2.
SP8 interacts with OsMYC3. (A) Interaction of SP8 with OsMYC3, but not with the OsMYC3 homologs OsMYC2 and OsMYC4, in the Y2H system. The full-length sequences of OsMYC2, OsMYC3, and OsMYC4 were amplified into pGADT7 (AD) and SP8 protein fused with pGBKT7 (BD) vector were cotransformed into the yeast strain AH109. Positive yeast transformants were selected on SD-L-T-H-Ade plates at 30 °C. Photos were taken after 3 days. (B) BiFC assays for potential interaction between SP8 and OsMYC3. SP8-cYFP was transiently coexpressed with OsMYC3-nYFP in N. benthamiana leaves and confocal imaged at 48 hpi. Agro-infiltration with SP8-cYFP and Gus-nYFP vector served as a negative control. (Scale bar, 20 μm.) (C) BiFC assays for potential interaction between SP8 and OsMYC3. SP8-cYFP was transiently coexpressed with OsMYC3-nYFP in N. benthamiana leaves and confocal imaged at 48 hpi. Agro-infiltration with SP8-cYFP and Gus-nYFP vector served as a negative control. (Scale bar, 20 μm.) (D) Co-IP assays to analyze the interactions among SP8 with OsMYC2 and OsMYC3 in vivo. Total proteins were incubated with FLAG beads and detected using anti-myc or anti-flag antibody. The sample coexpressing SP8-flag and GUS-myc was the negative control. (E) Co-IP assay confirms the interaction between OsMYC3 and the conserved NTP domain of SP8 in planta. SP8NTP-myc and OsMYC3-flag were coexpressed in N. benthamiana leaves, and total protein was incubated with FLAG beads and detected by anti-myc and anti-flag antibodies. Coinfiltration of SP8-myc and OsMYC3-flag was the positive control, while SP8NTP-myc cotransformed with flag-GFP served as a negative control. (F) Schematic representation of the deleted variations of OsMYC3 used in the Y2H assay. JID, JAZ interaction domain; TAD, transcriptional activation domain; bHLH, the basic helix–loop–helix domain responsible for homo-/hetero-dimerization. The right panel shows that OsMYC3TAD, but not OsMYC3jid or OsMYC3C, interacts with SP8.