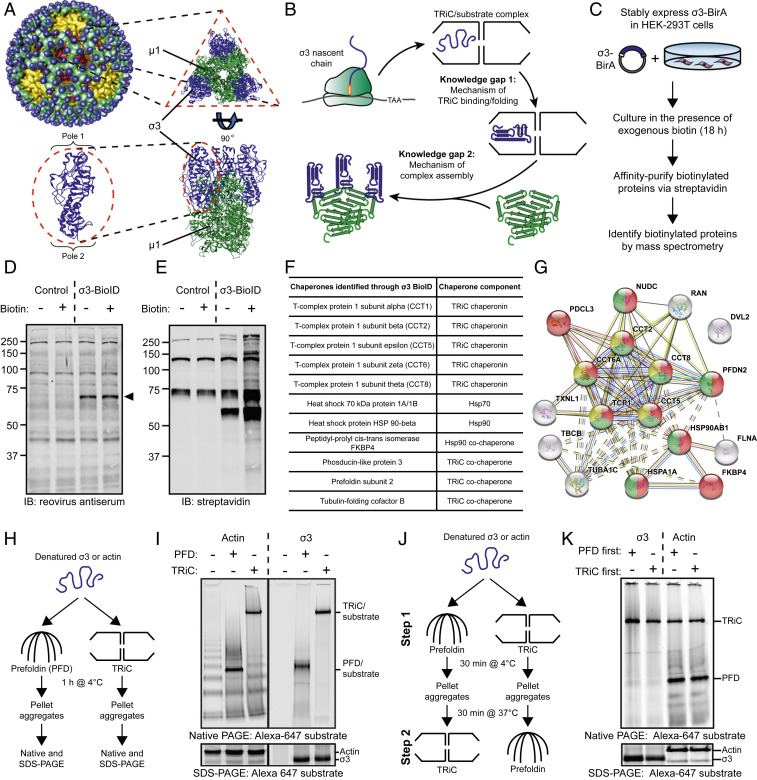
Fig. 1.
A chaperone network interacts with the reovirus σ3 major outer-capsid protein. (A) Structure of the reovirus particle (Upper Left) (44). Structure of the µ13σ33 outer-capsid heterohexamer and the σ3 monomer (Bottom Left); Protein Data Bank: 1JMU. (B) Model of the protein-folding pathway and knowledge gaps in the biogenesis of the reovirus outer capsid. (C) BioID workflow to identify host proteins that interact with the reovirus σ3 outer-capsid protein. (D and E) SDS-PAGE of control or σ3-BirA-expressing (σ3-BioID) HEK-293 cells incubated in the presence (+) or absence (−) of exogenous biotin for 18 h and immunoblotted with a reovirus-specific antiserum (D) or streptavidin (E). Arrowhead denotes σ3-BioID fusion protein. (F) Table of chaperones identified by σ3-BioID. (G) STRING analysis of host proteins identified by σ3-BioID. Yellow, TRiC subunits; red, protein-folding function; green, unfolded protein-binding function. (H) Workflow of PFD/TRiC/σ3-binding experiment. (I) Native- (Top) or SDS-PAGE (Bottom) of PFD/TRiC-binding experiment with σ3 or actin. (J) Workflow of two-step PFD/TRiC-binding experiment. (K) Native- (Top) or SDS-PAGE (Bottom) of two-step PFD/TRiC-binding experiment.