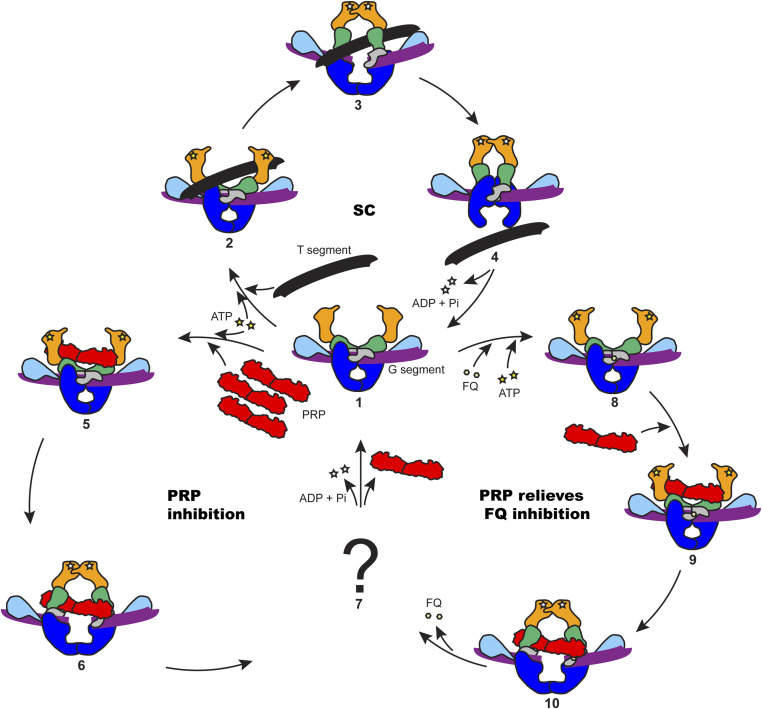
Fig. 8.
Proposed model for PRPs in the protection of DNA gyrase. Gyrase domains are colored as in Fig. 4. (1) Gyrase binds to a G segment, the N gate is open. (2) ATP is bound to GyrB and a T segment is presented to the ATPase domains of GyrB; the G segment is cleaved. (3) The presented T segment is captured by the GyrB clamp; ATP hydrolysis promotes the rotation of GyrB to open the DNA gate. (4) The T segment passes through the DNA gate and goes out through C (exit) gate; ADP and phosphate are released from GyrB. (5) Excess PRP is able to access the ATPase domains. (6) PRP is captured by the GyrB clamp and stimulates ATP hydrolysis, changing the conformation of GyrB to open the DNA gate. (7) The PRP is released in an unknown manner. (8) FQ stabilizes the gyrase–DNA cleavage complex. (9) PRP binds to the poisoned complex. (10) PRP activates the hydrolysis of ATP and changes the conformation of GyrB to open the DNA gate and dissociate the bound quinolones from the cleavage complex; the FQ is released from the complex. The supercoiling cycle (SC) contains 1, 2, 3, and 4. The PRP inhibition cycle contains 1, 5, 6, and 7. The PRP relieves FQ inhibition cycle contains 1, 8, 9, 10, and 7.