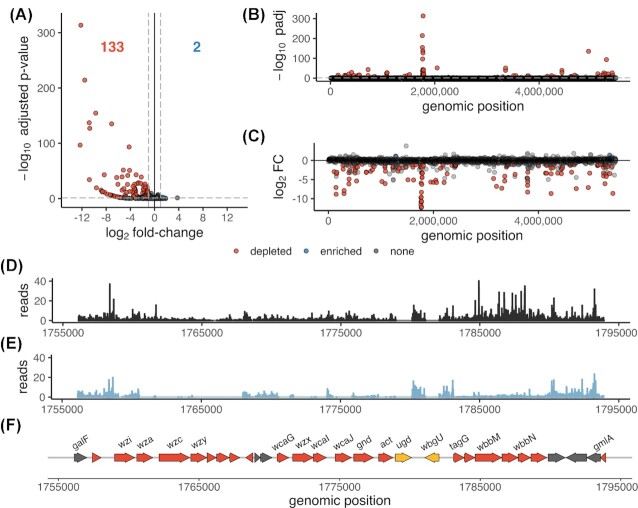
Figure 2.
TraDIS analysis identifies multiple potential K. pneumoniae fitness factors important during G. mellonella infections. (A), Volcano plot showing significantly less abundant genes 6 hours after infecting G. mellonella larvae in red and significantly higher abundant genes in blue. Non-significant genes are shown in grey. Genes were considered significant if they possessed a Benjamini–Hochberg corrected P-value below a threshold of 0.05 and an absolute log2 fold-change greater than 1. Essential and ambiguous-essential genes are removed from the analysis. (B and C), Manhattan plots with Benjamini–Hochberg corrected P-value and log2 fold-change on the y-axis, respectively. Abbreviations: padj, Benjamini-Hochberg corrected p-value; FC, fold-change. (D and E), Reads per nucleotide of the inoculum and at 6 hpi, respectively of the chromosomal region containing the K- and O-locus. For this graphical representation, triplicate TraDIS samples were combined and reads were normalised to the total number of reads in all three replicates. (F), Gene annotations of the Klebsiella K-locus (galF to wbgU) and O-locus (tagG to gmlA), encoding the polysaccharide capsule and lipopolysaccharide O antigen, respectively are coloured according to the TraDIS results at 6 hours post-infection; significantly depleted genes are shown in red, essential genes which were not included in the analysis in yellow and genes without significant changes in grey.