Figure 3:
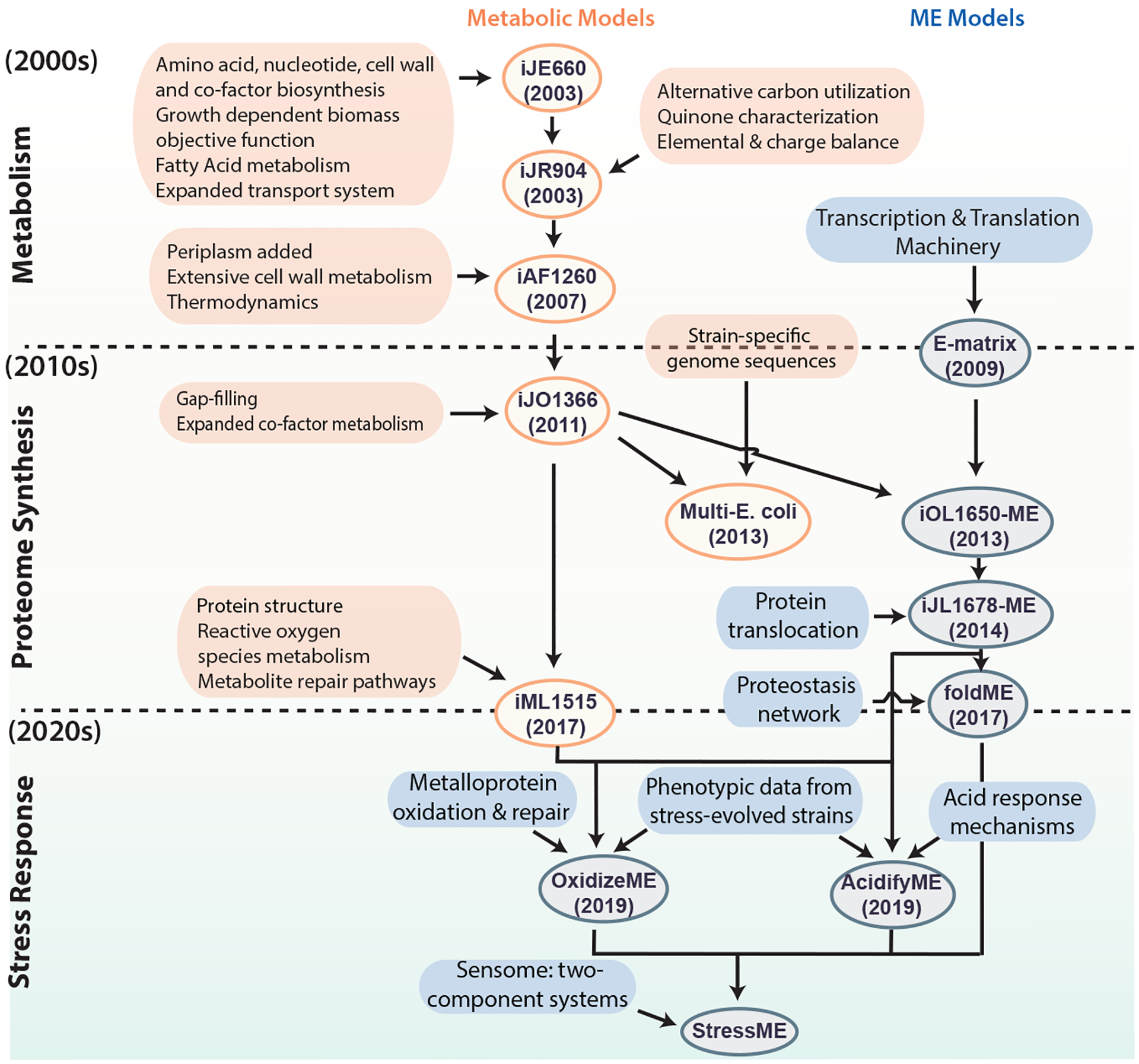
Historical development of Escherichia coli genome-scale models. Development of existing and potential future genome-scale models (both metabolic, shown in orange, and metabolic and macromolecular expression (ME) models shown in blue) of E. coli. The genome-scale metabolic model of E. coli first appeared in the early 2000s. An increasing scope of biological functions has been incorporated into the model, leading to various generations of the metabolic models as new discoveries were made. In the early 2010s, ME models that incorporate transcription and translation mechanisms emerged. Multiple efforts followed to improve and expand the ME model. Going into the 2020s, extensions of stress response modules have been added to ME models. Future directions involve incorporation of the sensome [G] to form the StressMe model, and the inclusion of toxins, biosynthetic gene clusters and cell cycle. Ovals indicate models, and boxes represent data incorporated to generate the models. According to the naming convention for network reconstructions, model names consist of an ‘i’ for in silico followed by the initials of the person(s) who built the model, and the number of open reading frames accounted for in the reconstruction.
