Figure 4:
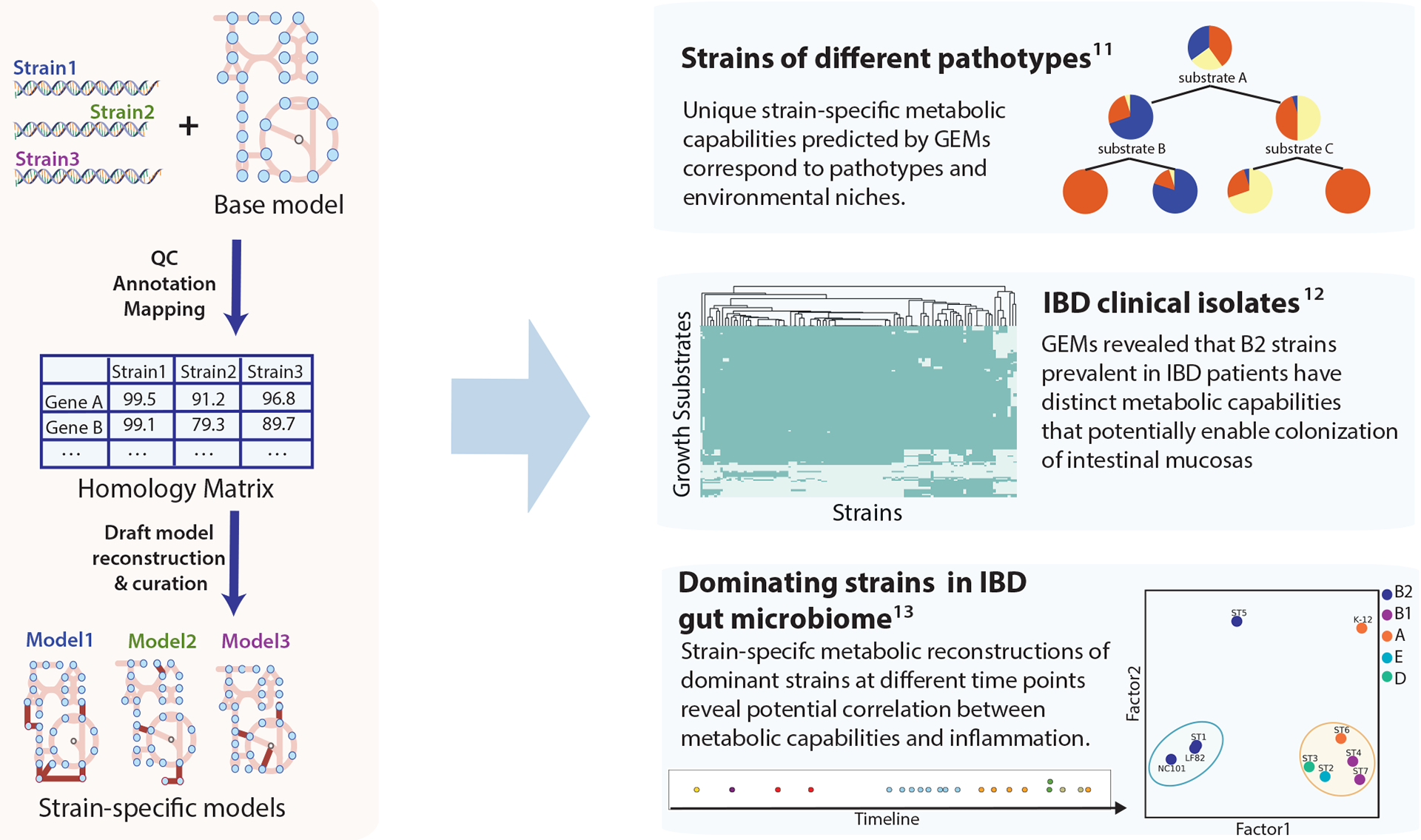
Generation of strain-specific Escherichia coli genome-scale models and their application to multi-strain studies. Strain-specific models were generated from genome sequences of strains of interest and a curated reference model. The annotated genome sequences of target strains are mapped to the reference genome sequence to generate the homology matrix that delineates the gene sequence similarity across strains. The homology matrix can be used to create draft models of target strains. These models can then be finalized by manual curation. Strain-specific models were used to reveal variation in metabolic capabilities across different pathotypes, as illustrated in three studies shown on the right. The first multi-strain study of E. coli genome-scale models (GEMs) found metabolic capabilities predicted by GEMs correspond to pathotype and environment. In the second study, comparison of GEMs constructed for inflammatory bowel disease (IBD) clinical isolates suggested the possible link between metabolic functions of B2 strains and their prevalence in individuals with IBD. Lastly, GEMs of dominant strains in an individual with IBD revealed the potential correlation between metabolism and inflammation91–93. Panel ‘Strains of different pathotypes’ adapted from ref. 91. Panel ‘IBD clinical isolates’ adapted from ref. 92. Panel ‘Dominating strains in IBD gut microbiome’ adapted from ref. 93.
