Figure 5:
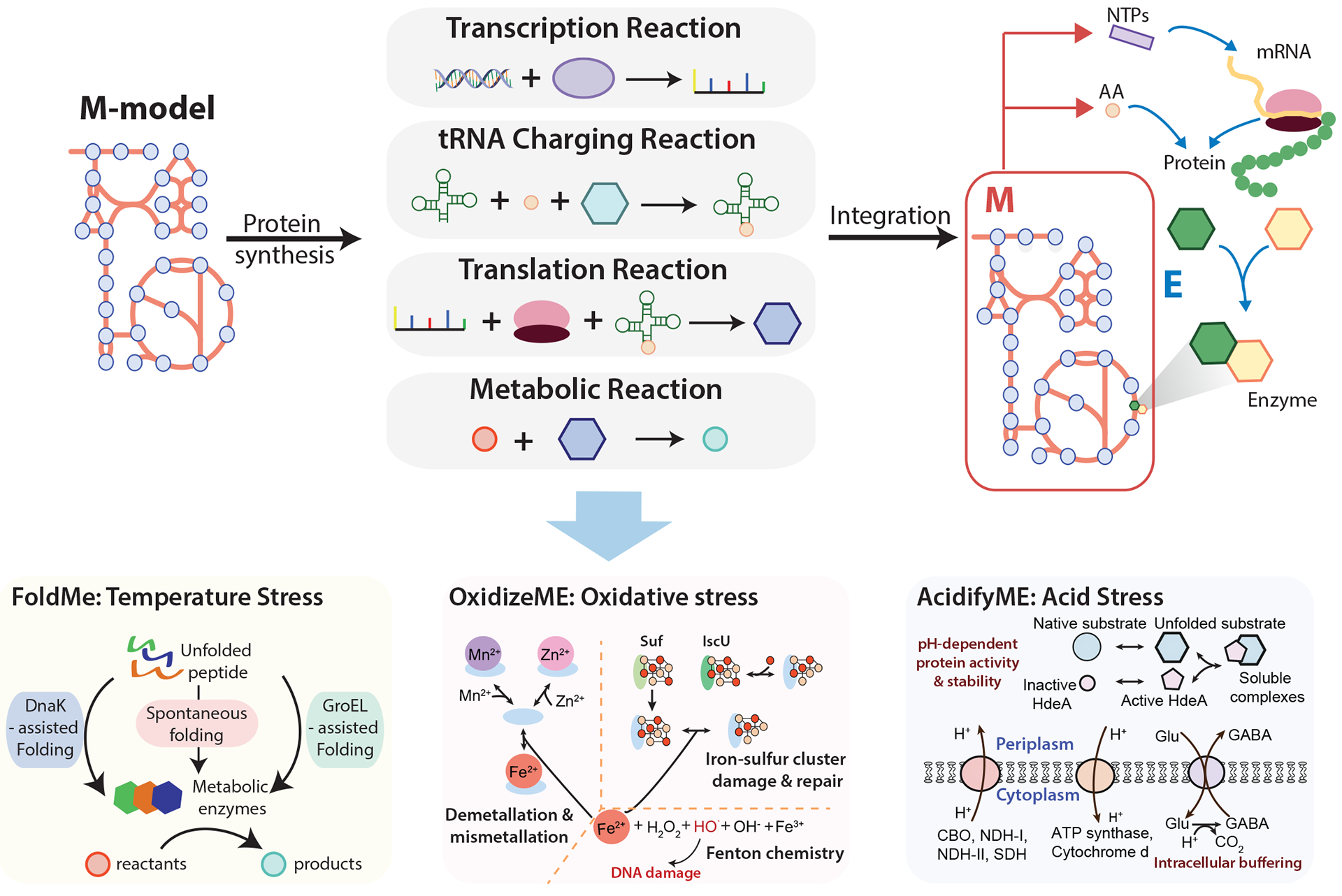
General formulation of a metabolic and macromolecular expression model and its application to the study of stress response. Metabolic and macromolecular expression (ME) models are generated through the integration of M models and protein synthesis pathways including transcription, tRNA charging, and translation. Therefore, the ME model describes the biosynthesis of proteins and their roles in catalyzing the metabolic reactions. Stress-specific response mechanisms are integrated with the E. coli ME model to produce stress-specific ME models: FoldME, OxidizeME, and AcidifyME. FoldME models respond to temperature stress through the incorporation of chaperone-mediated (GroEL or DnaK) or spontaneous folding pathways. OxidizeME simulates the response to oxidative stress through the inclusion of oxidation and demetallation in metalloproteins, iron-sulfur cluster cofactor damage and repair, and DNA damage. AcidifyME models the mechanisms related to acid stress, including pH-dependent protein activity and stability, membrane composition, and intracellular buffering. CBO, cytochrome bo terminal oxidase; NDH-I, NADH dehydrogenase I; NDH-II, NADH dehydrogenase II; SDH, succinate dehydrogenase; Glu, glutamate; GABA, gamma-aminobutyric acid; NTPs, nucleoside triphosphates. FoldME panel adapted from ref. 127. OxidizeME panel adapted from ref. 129.
