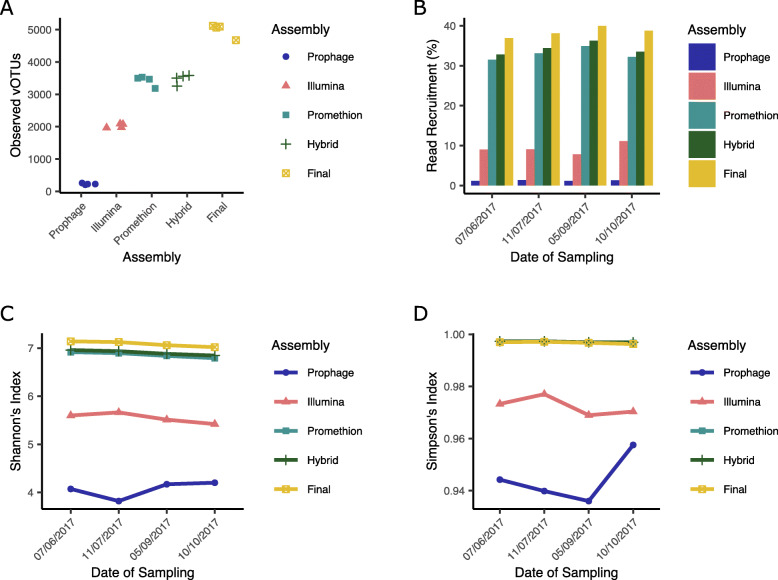
Fig. 2.
Abundance and diversity of vOTUs in different assemblies. a Number of vOTUs observed in each sample obtained from normalised read counts. The hybrid assembly is the combination of both Illumina and PromethION reads. Prophage were predicted from a bacterial metagenome from the same sample. Final assembly was combination of Illumina, hybrid and identified active prophage where were dereplicated at 95% ANI. b Read recruitment over time for the different assemblies. c Shanon’s ⍺-diversity from different assemblies for each sampling point. d Simpson’s ⍺-diversity assemblies for each sampling point