Fig. 3.
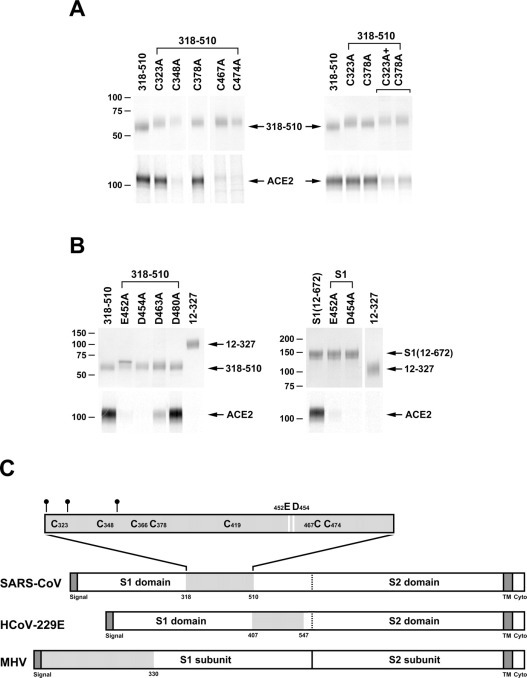
Analysis of point mutations of S1-Ig and the 318–510 variant.A, the 318–510 S1-Ig truncation variant, or variants thereof in which each of seven cysteines was altered individually to alanine, were analyzed as described in the legend to Fig. 1. Variants in which cysteine 366 or 419 was altered to alanine did not express and were not further analyzed. A variant containing alterations of both cysteines 323 and 378 was also analyzed (right panel). B, 318–510 variants (left panel) or S1-Ig variants (right panel) in which glutamic acid 452 or aspartic acids 454, 463, or 480 were altered individually to alanine were analyzed as in Fig. 1. C, representation of the S proteins of SARS-CoV, HCoV-229E, and MHV, aligned by their S2 domains. Dark gray indicates leader and transmembrane sequences. Light gray indicates receptor-binding domain. The receptor-binding domain of SARS-CoV is shown with N-glycosylation sites (small circles) and cysteines indicated. Residues that make a substantial contribution to ACE2 association (glutamic acid 452 and aspartic acid 454) are shown as white bars.
