Fig. 7. IL-7-regulated transcripts in Crk/CrkL conditionally-deficient pro-B cells.
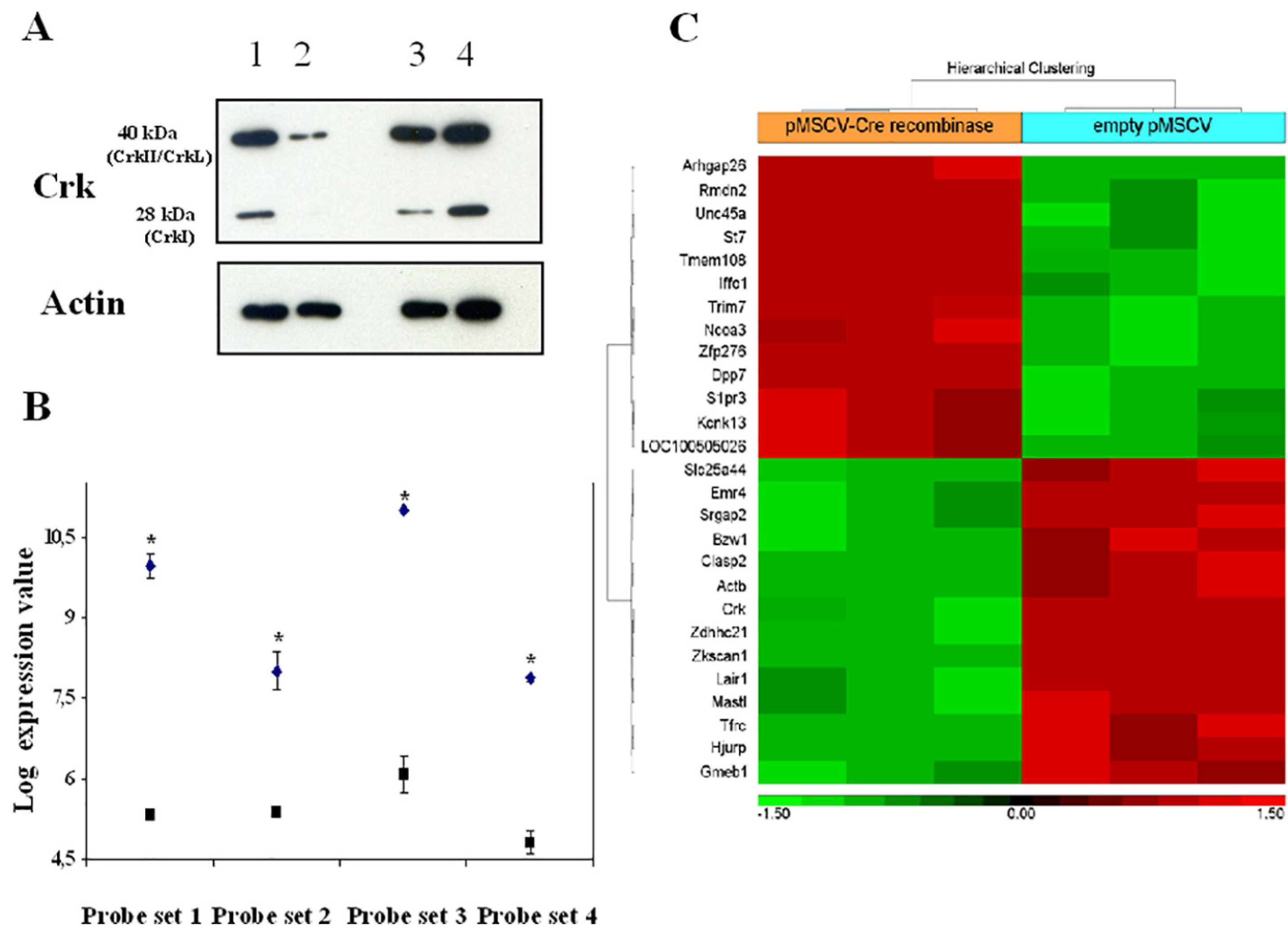
Cells from mice bearing loxP-flanked Crk/CrkL alleles cultured with IL-7 for 67 h were transduced with the retroviral vector pMSCV-Cre recombinase-IRES-GFP or with the control empty vector (pMSCV-IRES-GFP). A, Bone marrow cells enriched in pro-B cells: Western blot analysis with anti-Crk moAb. Lane 1: no vector; lane 2: pMSCV-Cre recombinase-IRES-GFP; lane 3: no vector; lane 4: empty vector pMSCV-IRES-GFP; B, RNA purified from pro-B cells was analysed by microarray as described in Materials and Methods. The 4 probe sets (1,416,201, 1,460,176 and 1,448,248) at FDR P-val < 0.05 (*), and 1,425,855 at nominal P-val < 0.05 (*) recognized homologous regions of Crk and CrkL RNA. Closed squares: cells transduced with pMSCV-Cre recombinase-IRES-GFP; closed diamonds: cells transduced cells with the control empty vector pMSCV-IRES-GFP. Representative of 3 experiments; C, Hyerarchical clustering of genes that differentially showed upregulation (> 2 fold-increase, red) or downregulation (<−2 fold-decrease, green). Bone marrow cells enriched in pro-B cells from C57BL/6 mice bearing loxP-flanked Crk/CrkL alleles cultured with IL-7 for 67 h were transduced with the retroviral vector pMSCV-Cre recombinase-IRES-GFP or with the control vector pMSCV-IRES-GFP. RNA from purified pro-B cells was analysed as described in Materials and Methods. Representative of 3 experiments.
