Figure 2.
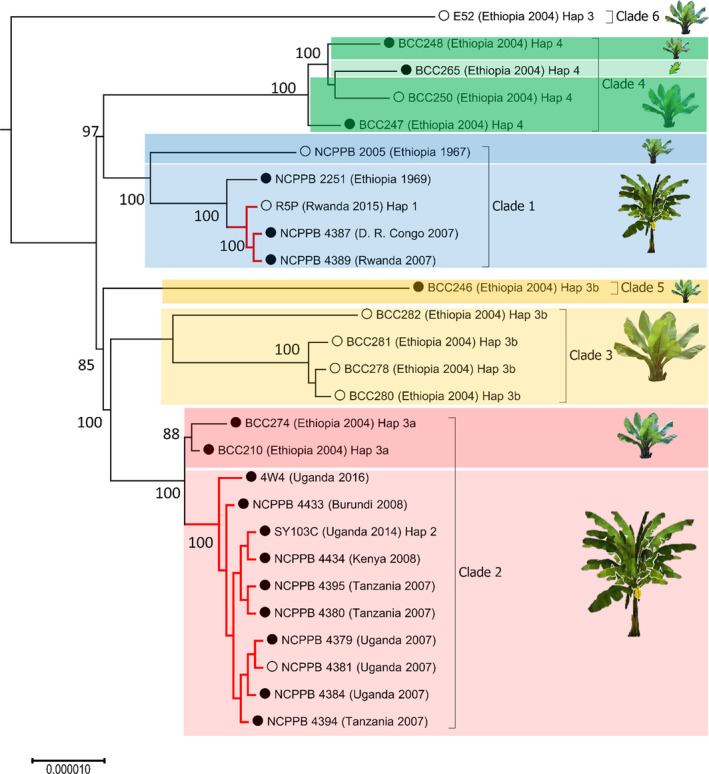
Genome‐based phylogenetic analysis of Xanthomonas vasicola pv. musacearum (Xvm) isolates. Phylogenetic tree analysis was performed on FastQ files using REALPHY (Bertels et al., 2014), applying the RaxML algorithm (Stamatakis, 2014) for tree constructing (NCPPB 4379 as reference genome). R5P, 4W4, SY103C, E52, and all BCC genomes were newly sequenced in the present study, while other Xvm genome sequences were published previously (Studholme et al., 2010; Wasukira et al., 2012). The tree was rooted on the genome sequence of the reference X. vasicola pv. holcicola NCPPB 2417 (not shown). The plasmid pXCM49 is present in the closed circle genomes, while absent from the open circle genomes. Red branches indicate genomes sampled out of Ethiopia. Node robustness is indicated by bootstrap values (percentages from 500 trials). All Ethiopian strains were isolated from enset, except NCPPB 2251 (banana) and BCC265 (maize). All other strains were isolated from banana
