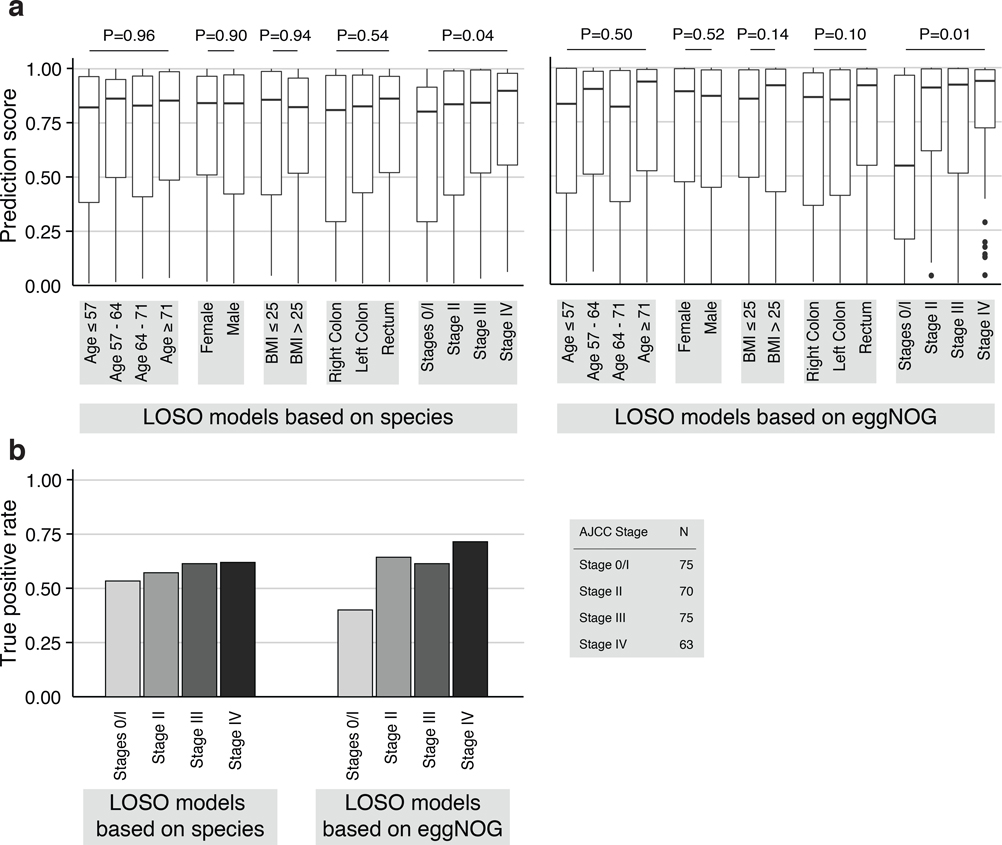
Extended Data Figure 7: Analysis of leave-one-study-out models for prediction bias.
(a) To examine whether species and gene-family-level classification models are confounded, i.e. biased towards certain patient subgroups, prediction scores from leave-one-study-out models are plotted broken down into strata for each clinical parameter (e.g. female and male for sex). Prediction bias for each variable was tested by two-sided Wilcoxon (for sex and BMI) or Kruskal-Wallis (all others) tests while blocking for study as confounder (n=575 independent observations). Boxes denote interquartile ranges (IQR) with the median as horizontal black line and whiskers extending up to the most extreme point within 1.5-fold IQR. A significant difference in prediction score was detected only for CRC stage. This stage-bias is more pronounced for gene-family then for species models. (b) To examine CRC stage bias further the barplots show the true positive rate (TPR) corresponding to an overall 10% false positive rate (see also Fig. 3c) for the different CRC stages displaying slightly higher classification sensitivity for late stage CRC for both species and gene-family models.