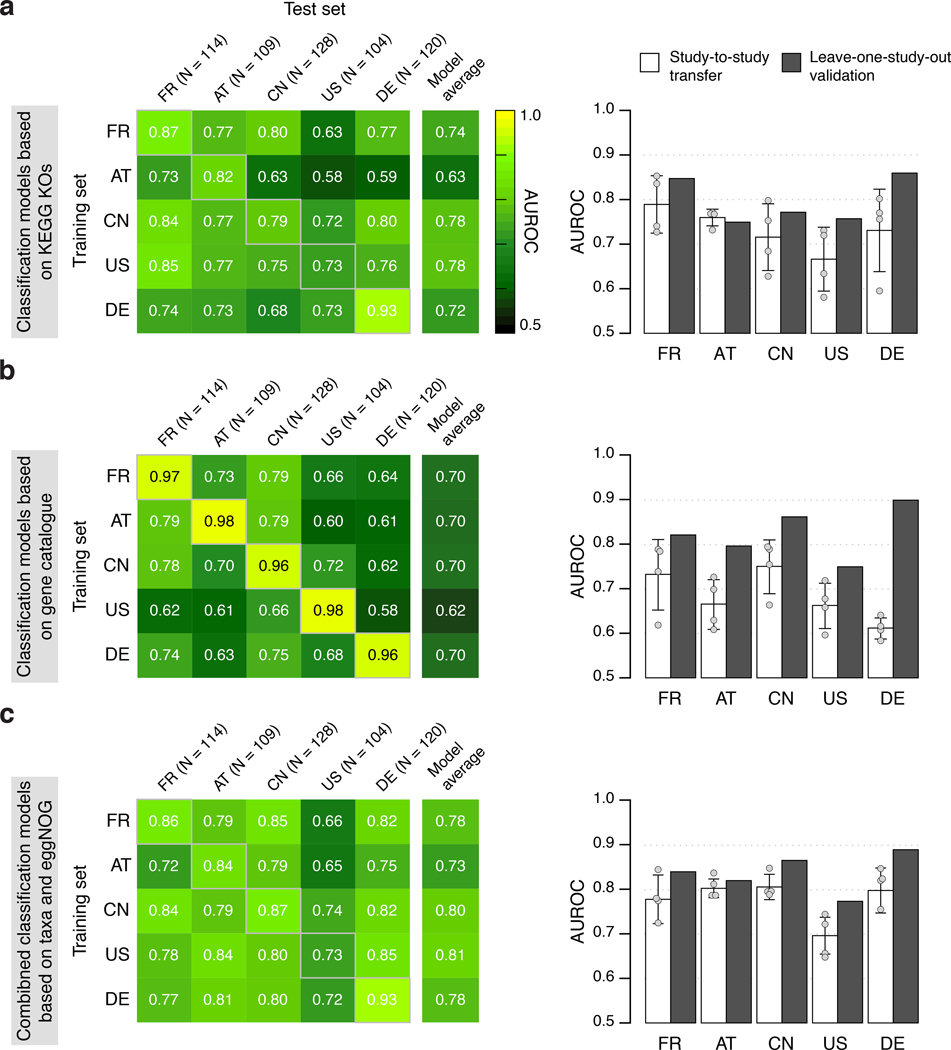
Extended Data Figure 8: Cross-study performance of statistical models based on KEGG KO abundances, single-gene abundances from the metagenomic gene catalogue (IGC), and the combination of taxonomic and eggNOG abundance profiles.
CRC classification accuracy resulting from cross validation within each study (gray boxed along diagonal) and study-to-study model transfer (external validations off diagonal) as measured by AUROC for classification models trained on KEGG KO (a), models based on the gene catalogue (b), and models based on the combination of taxonomic and eggNOG abundance profiles (c) (see Methods for details on statistical modeling workflows). The last column depicts the average AUROC across external validations. The barplots on the right show that the classification accuracy on a held-out study improves if data from all other studies are combined for training (leave-one-study-out, LOSO validation) relative to the mean of models trained on data from a single study (study-to-study transfer, n=4, error bars show standard deviation) consistently across different types of input data.