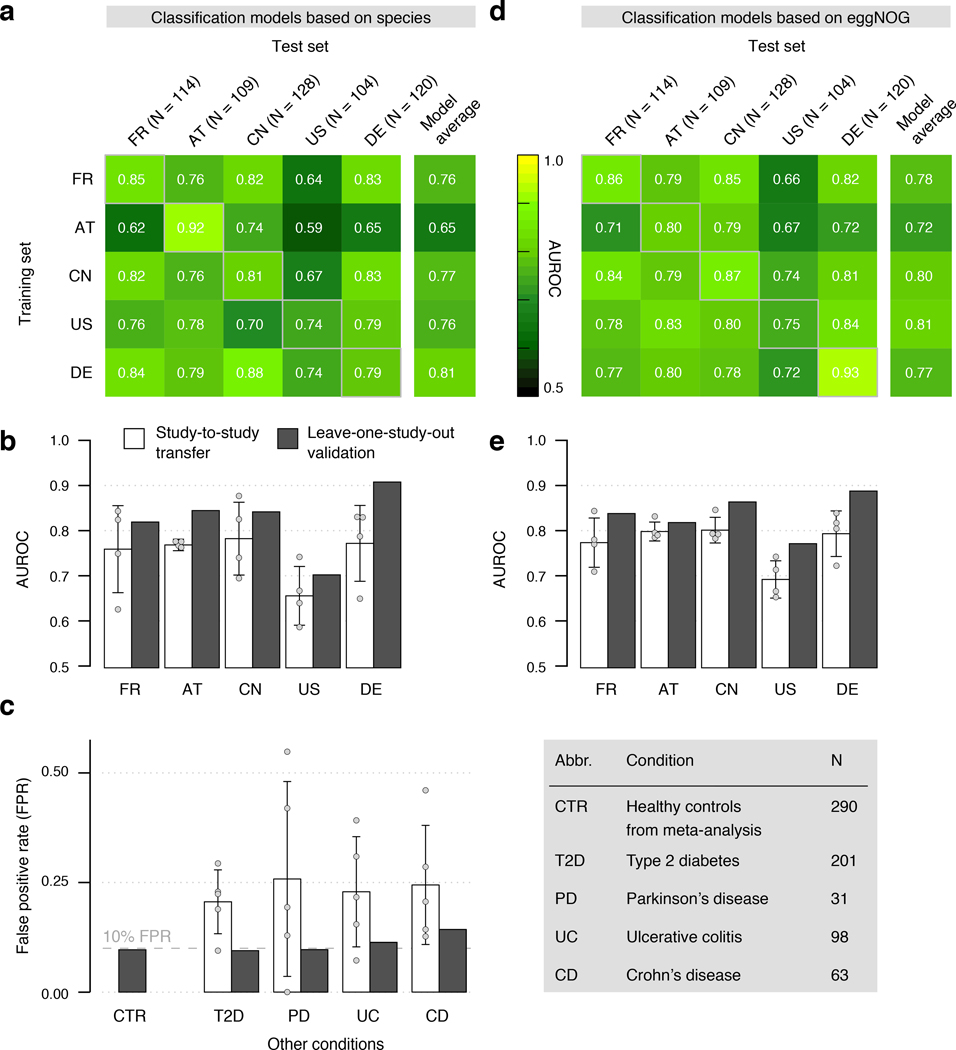
Figure 3. Both taxonomic and functional metagenomic classification models generalize across studies in particular when trained on data from multiple studies.
CRC classification accuracy resulting from cross validation within each study (gray boxes along diagonal) and study-to-study model transfer (external validations off diagonal) as measured by AUROC for classifiers trained on (a) species and (d) eggNOG gene family abundance profiles. The last column depicts the average AUROC across external validations. Classification accuracy, as evaluated by AUROC on a held-out study, improves if taxonomic (b) or functional (e) data from all other studies are combined for training (leave-one-study-out, LOSO validation) relative to models trained on data from a single study (study-to-study transfer, average and standard deviation shown). Bar height for study-to-study transfer corresponds to the average of four classifiers (error bars indicate standard deviation, n=4). (c) Combining training data across studies substantially improves CRC specificity of the (LOSO) classification models relative to models trained on data from a single study (depicted by bar color, as in (c) and (d)) as assessed by the false positive rate (FPR) on fecal samples from patients with other conditions (see legend). Bar height for study-to-study transfer corresponds to the average FPR across classifiers (n=5) with error bars indicating the standard deviation of FPR values observed.