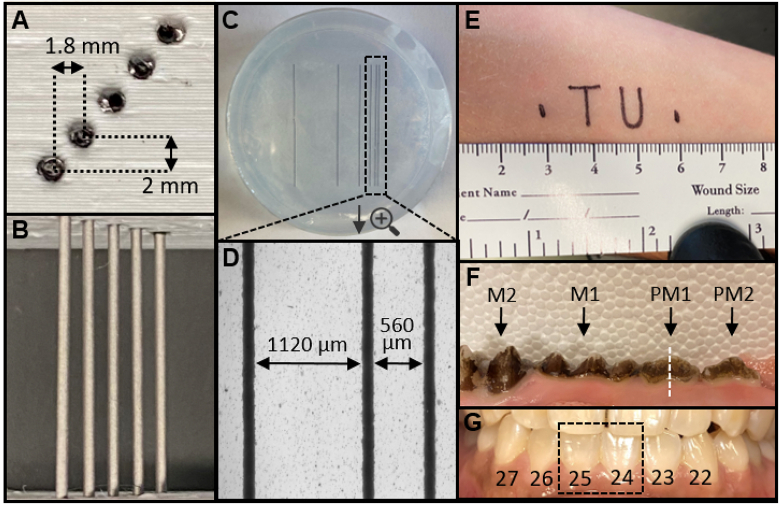
Fig. 2.
Imaging phantoms and targets. (a) Photographic cross-section of depth phantom 3D-printed from PLA. The holes are 1-mm diameter and filled with 0.8 mm graphite pencil lead. The lateral spacing is 1.8 mm and the axial spacing is 2 mm. (b) Top-down photograph of depth phantom. (c) Top-down photograph of lateral resolution phantom, consisting of inkjet-printed lines on transparent film embedded in 1% agar within a 5.5-cm petri dish. The spacing between each line (width = 130 µm) doubles sequentially and was measured with brightfield microscopy. From right to left, d1 = 560 µm, d2 = 1120 µm, d3 = 2240 µm, d4 = 4480 µm, d5 = 8960 µm. (d) Spacing between the right-most (closest) lines is shown with brightfield microscopy. (e) Text printed with permanent marker on the supinated forearm to evaluate algorithm performance for a non-linear target. The total feature length was 3 cm. (f) Photograph of the ex vivo swine maxilla with teeth labeled: M2 (2nd molar), M1 (1st molar), PM1 (1st premolar), PM2 (2nd premolar). The white dashed line indicates the sagittal imaging plane generated from transverse B-mode images. (g) Photograph of the teeth and gingiva imaged in a healthy human subject labeled with the universal numbering system—the dashed box shows teeth imaged for deshaking.