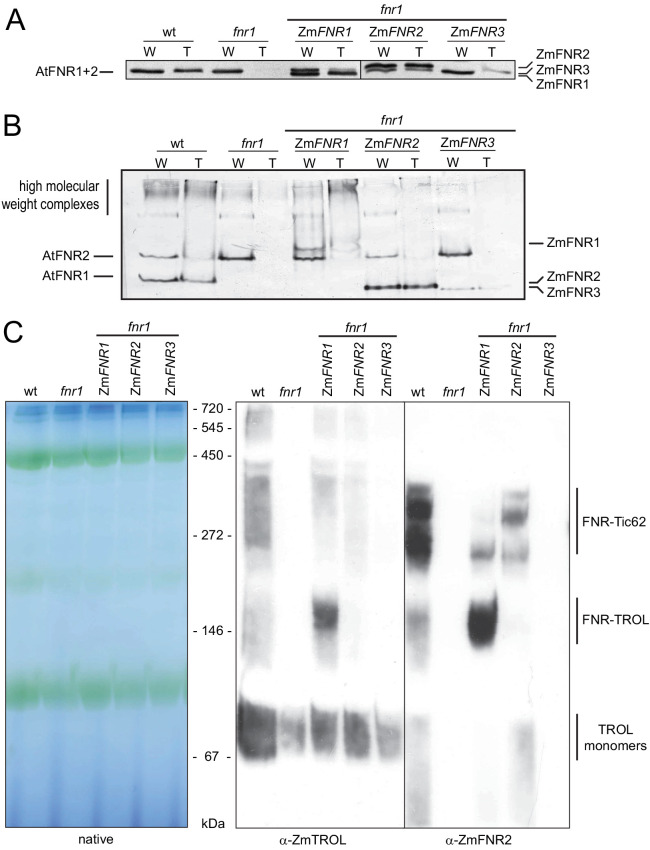
Figure 2. Arabidopsis plants with variable FNR:membrane tether interactions.
Leaf extracts of Arabidopsis wt, fnr1, and fnr1 mutants transformed to express genes for the maize FNR proteins ZmFNR1, ZmFNR2, and ZmFNR3 in the fnr1 background were separated into soluble and insoluble fractions. These were designated loose (L) and tight (T) membrane bound FNR fractions, based on the analysis in Figure 1. Samples were subjected to (A) SDS-PAGE (25 µg protein prior to separation of L and T fractions) or (B) native PAGE (20 µg protein prior to separation of L and T fractions) before immunoblotting, challenge with antisera against FNR, then detection with alkaline phosphatase. Migration positions of Arabidopsis (At) FNRs and maize (Zm) FNRs are indicated to the left and right, respectively. (C) Recruitment of maize FNR proteins into specific Arabidopsis thylakoid membrane complexes. Chloroplasts were isolated from the same genotypes used in (A) and (B). Thylakoid membranes were solubilised with DDM and subjected to blue native-PAGE (BNP). Samples were loaded on an equal chlorophyll basis, with 3.2 μg per lane, western blotted and challenged with antisera raised against FNR (rabbit) and TROL (guinea pig) before visualisation using secondary antisera conjugated to horseradish peroxidase (chemiluminescence) and alkaline phosphatase respectively. Positions of molecular mass markers are indicated between the gels and the blots in kDa.