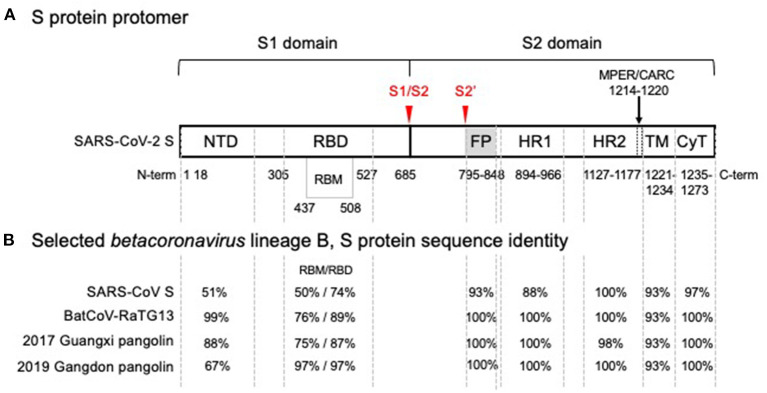
Figure 1.
SARS-CoV-2 spike glycoprotein monomer representation showing (A) functional domains and (B) comparison of amino acid sequence identity with SARS-CoV and related isolates in the wild. (A) Functional domain S1 mediates binding of the receptor binding domain (RBD) to the angiotensin converting enzyme 2 (ACE2), the host cell receptor that is specifically recognized by the receptor binding motif (RBM) interface, is cleaved (S1/S2) and shed. Shedding exposes the S2 domain. Cleavage at S2' triggers spike trimer activation, release of the fusion peptide (FP), heptad repeat 1 (HR1) and heptad repeat 2 (HR2); the membrane proximal external region (MPER) is sometimes considered part of HR2 and with a cholesterol recognition/interaction amino acid consensus (CARC) sequence, potentially participating in membrane lipid fusion. The transmembrane domain (TM) and a short cytoplasmic tail (CyT) are indicated. Cleavage sites that drive host-cell infection are shown in red. (B) Phylogenetic analysis of SARS-CoV-2 domain sequences identity among SARS-CoV-2, bat and pangolin's isolates, and SARS-CoV are tabulated for each domain. A high degree of sequence identity for the RBD in SARS-CoV correlates well with ACE2 receptor recognition. However, the RBM-ACE2 interface is 50% identical and may account for the increased ACE2 binding affinity. Sequence identities are high for all functional segments of S2, the Type I metastable domain that induces viral-host cell membrane fusion, suggesting an optimum sequence-structure-function relationship. Metastability is a functional requirement, allowing these proteins to refold into a lower energy conformation while transferring the difference in energy to catalyze the membrane fusion reaction. Structural studies have shown that stable immunogens presenting the same antigenic sites as the labile wild-type proteins efficiently elicit potent neutralizing antibodies. In the alternative endosomal pathway that is apparently mediated by NTD, the fusion machine of S2 is equally required to infect the host cells (9, 10). Furthermore, detrimental amino acid substitutions found in the S2 domain of Hepatitis Mouse Virus, but not in SARS-CoV-2, affected the post fusion conformational stability, explaining a reported reduction of S-mediated membrane fusion (11).